Homologous Recombination-Enhancing Factors Identified by Comparative Transcriptomic Analyses of Pluripotent Stem Cell of Human and Common Marmoset
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Vector Construction
2.3. Cell Culture, Transfection, and Drug Selection
2.4. Genotyping
2.5. Quantitative Reverse-Transcription PCR (qPCR)
2.6. RNA-Seq Analysis
2.7. Western Blotting
2.8. Statistical Analysis
3. Results
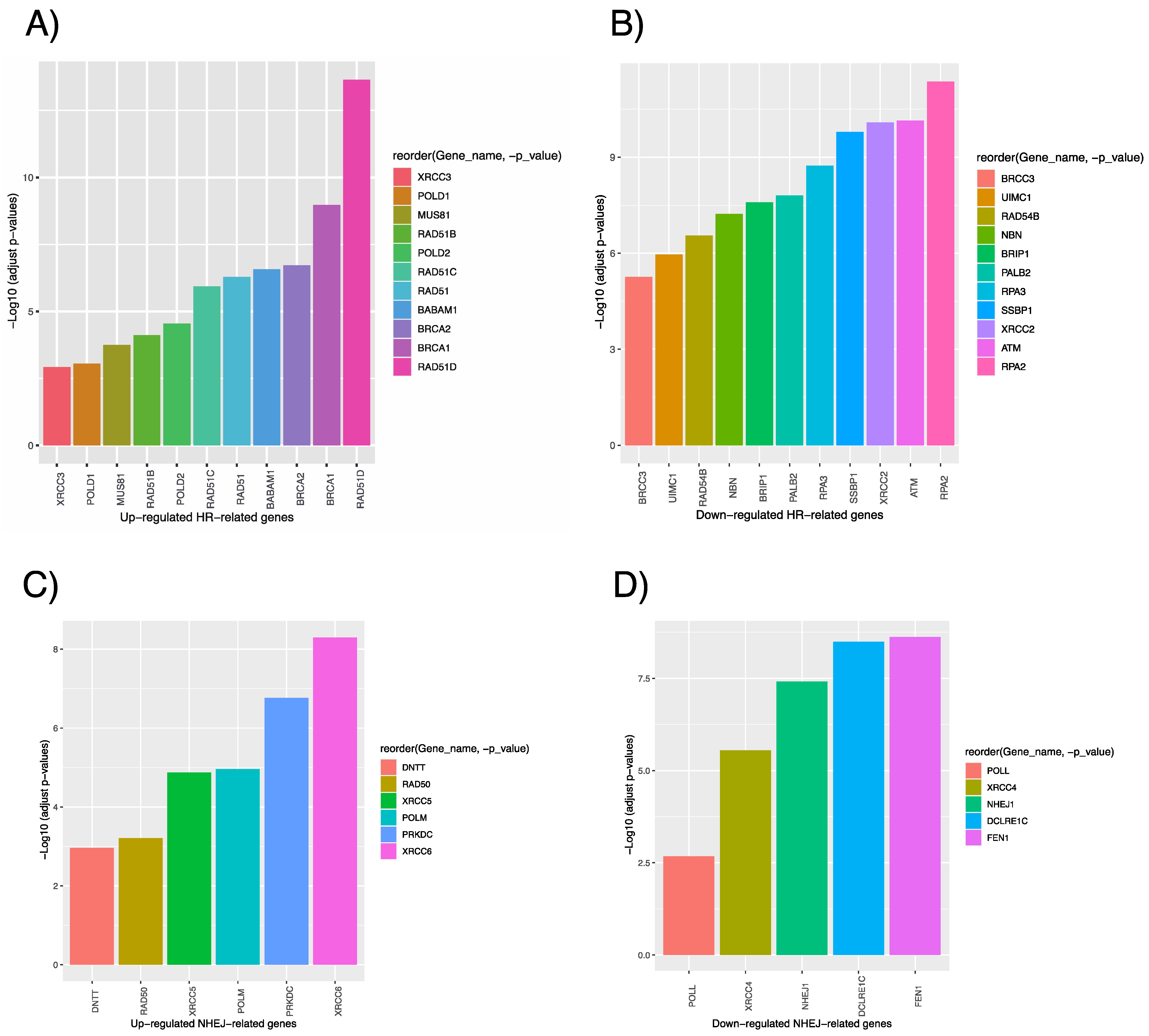
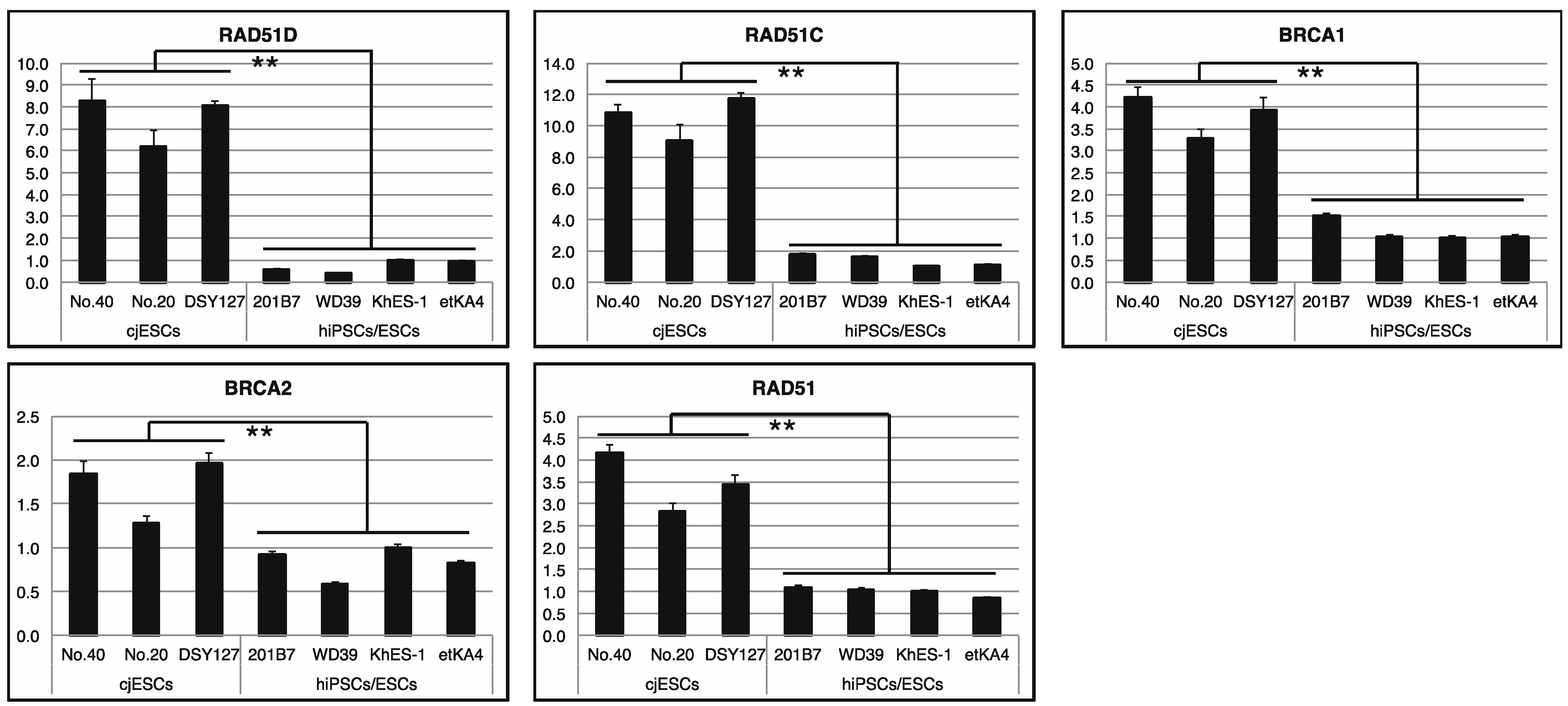
3.1. Comparative Transcriptomic Analysis of Human and Marmoset PSCs
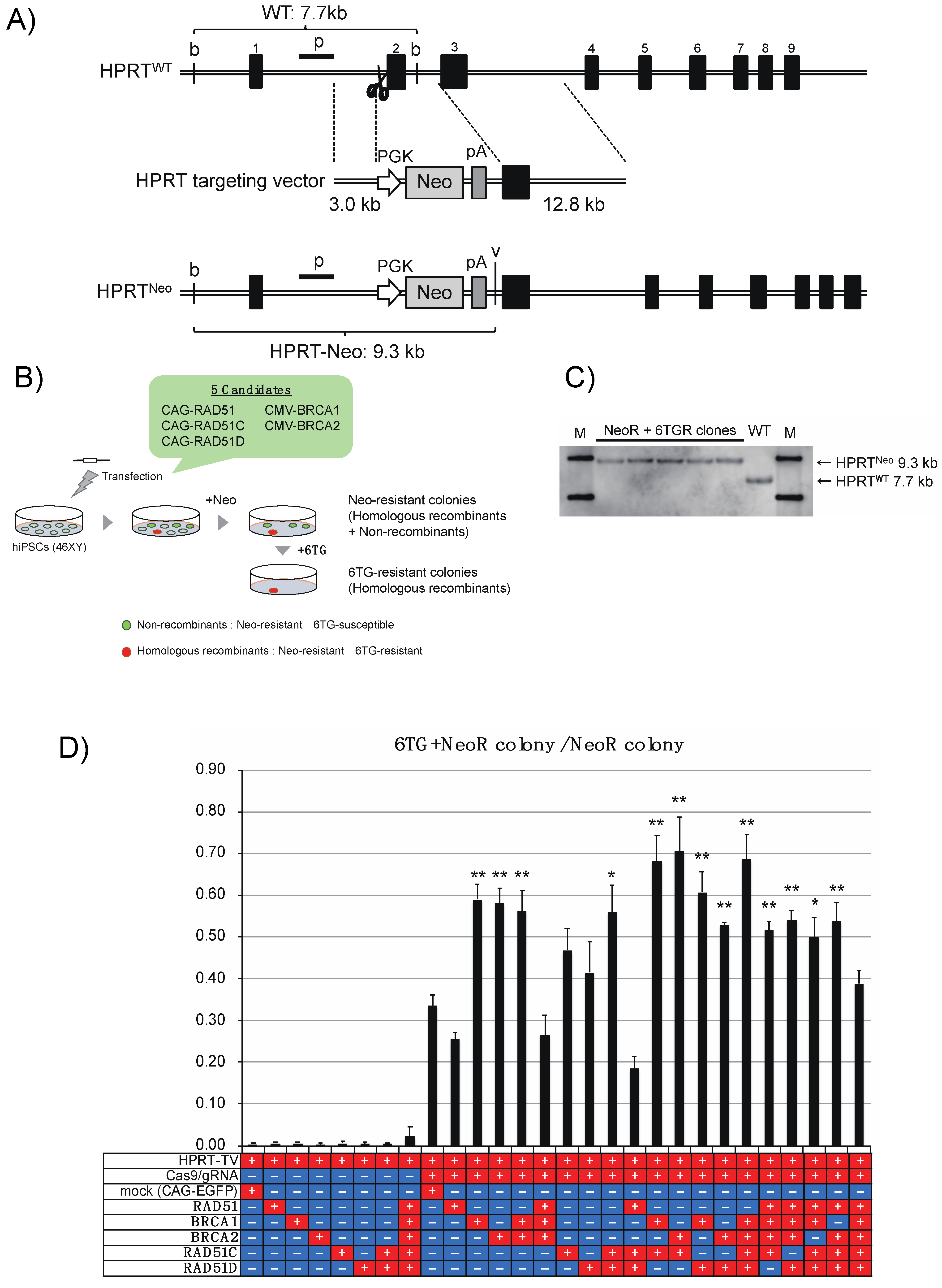
3.2. Enhancement of the HR and RI Ratio with CRISPR-Cas9 in hiPSCs by Overexpression of the Defined Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [Green Version]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Buerstedde, J.M.; Takeda, S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 1991, 67, 179–188. [Google Scholar] [CrossRef]
- Sung, P.; Klein, H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 739–750. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef] [Green Version]
- Pinder, J.; Salsman, J.; Dellaire, G. Nuclear domain ‘knock-in‘ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015, 43, 9379–9392. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kuhn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Tran, N.T.; Bashir, S.; Li, X.; Rossius, J.; Chu, V.T.; Rajewsky, K.; Kuhn, R. Enhancement of Precise Gene Editing by the Association of Cas9 With Homologous Recombination Factors. Front. Genet. 2019, 10, 365. [Google Scholar] [CrossRef] [Green Version]
- Shao, S.; Ren, C.; Liu, Z.; Bai, Y.; Chen, Z.; Wei, Z.; Wang, X.; Zhang, Z.; Xu, K. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 2017, 92, 43–52. [Google Scholar] [CrossRef]
- Charpentier, M.; Khedher, A.H.Y.; Menoret, S.; Brion, A.; Lamribet, K.; Dardillac, E.; Boix, C.; Perrouault, L.; Tesson, L.; Geny, S.; et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018, 9, 1133. [Google Scholar] [CrossRef] [PubMed]
- Jayavaradhan, R.; Pillis, D.M.; Goodman, M.; Zhang, F.; Zhang, Y.; Andreassen, P.R.; Malik, P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019, 10, 2866. [Google Scholar] [CrossRef] [Green Version]
- Nambiar, T.S.; Billon, P.; Diedenhofen, G.; Hayward, S.B.; Taglialatela, A.; Cai, K.; Huang, J.W.; Leuzzi, G.; Cuella-Martin, R.; Palacios, A.; et al. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat. Commun. 2019, 10, 3395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, H.; Yamanaka, S. iPS cell technologies: Significance and applications to CNS regeneration and disease. Mol. Brain 2014, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Okano, H.; Sipp, D. New trends in cellular therapy. Development 2020, 147, dev192567. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell 2012, 10, 678–684. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, S. Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef]
- Yoshimatsu, S.; Sone, T.; Nakajima, M.; Sato, T.; Okochi, R.; Ishikawa, M.; Nakamura, M.; Sasaki, E.; Shiozawa, S.; Okano, H. A versatile toolbox for knock-in gene targeting based on the Multisite Gateway technology. PLoS ONE 2019, 14, e0221164. [Google Scholar] [CrossRef]
- Sasaki, K.; Yokobayashi, S.; Nakamura, T.; Okamoto, I.; Yabuta, Y.; Kurimoto, K.; Ohta, H.; Moritoki, Y.; Iwatani, C.; Tsuchiya, H.; et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 2015, 17, 178–194. [Google Scholar] [CrossRef] [Green Version]
- Irie, N.; Weinberger, L.; Tang, W.W.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef] [Green Version]
- Sybirna, A.; Tang, W.W.C.; Pierson Smela, M.; Dietmann, S.; Gruhn, W.H.; Brosh, R.; Surani, M.A. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020, 11, 1282. [Google Scholar] [CrossRef]
- Zwaka, T.P.; Thomson, J.A. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003, 21, 319–321. [Google Scholar] [CrossRef]
- Nakamura, M.; Shiozawa, S.; Tsuboi, D.; Amano, M.; Watanabe, H.; Maeda, S.; Kimura, T.; Yoshimatsu, S.; Kisa, F.; Karch, C.M.; et al. Pathological Progression Induced by the Frontotemporal Dementia-Associated R406W Tau Mutation in Patient-Derived iPSCs. Stem Cell Rep. 2019, 13, 684–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichiyanagi, N.; Fujimori, K.; Yano, M.; Ishihara-Fujisaki, C.; Sone, T.; Akiyama, T.; Okada, Y.; Akamatsu, W.; Matsumoto, T.; Ishikawa, M.; et al. Establishment of In Vitro FUS-Associated Familial Amyotrophic Lateral Sclerosis Model Using Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2016, 6, 496–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimori, K.; Ishikawa, M.; Otomo, A.; Atsuta, N.; Nakamura, R.; Akiyama, T.; Hadano, S.; Aoki, M.; Saya, H.; Sobue, G.; et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 2018, 24, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, F.K.; Okamoto, S.; Mitsui, J.; Sone, T.; Ishikawa, M.; Yamamoto, Y.; Kanegae, Y.; Nakatake, Y.; Imaizumi, K.; Ishiura, H.; et al. The pathogenesis linked to coenzyme Q10 insufficiency in iPSC-derived neurons from patients with multiple-system atrophy. Sci. Rep. 2018, 8, 14215. [Google Scholar] [CrossRef] [PubMed]
- Simara, P.; Motl, J.A.; Kaufman, D.S. Pluripotent stem cells and gene therapy. Transl. Res. 2013, 161, 284–292. [Google Scholar] [CrossRef] [Green Version]
- Ou, Z.; Niu, X.; He, W.; Chen, Y.; Song, B.; Xian, Y.; Fan, D.; Tang, D.; Sun, X. The Combination of CRISPR/Cas9 and iPSC Technologies in the Gene Therapy of Human beta-thalassemia in Mice. Sci. Rep. 2016, 6, 32463. [Google Scholar] [CrossRef] [Green Version]
- Takayama, K.; Igai, K.; Hagihara, Y.; Hashimoto, R.; Hanawa, M.; Sakuma, T.; Tachibana, M.; Sakurai, F.; Yamamoto, T.; Mizuguchi, H. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 2017, 45, 5198–5207. [Google Scholar] [CrossRef] [PubMed]
- Hockemeyer, D.; Soldner, F.; Beard, C.; Gao, Q.; Mitalipova, M.; DeKelver, R.C.; Katibah, G.E.; Amora, R.; Boydston, E.A.; Zeitler, B.; et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009, 27, 851–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.J.; Zhang, L.; Santiago, Y.; Miller, J.C.; et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011, 29, 731–734. [Google Scholar] [CrossRef] [Green Version]
- Sebastiano, V.; Maeder, M.L.; Angstman, J.F.; Haddad, B.; Khayter, C.; Yeo, D.T.; Goodwin, M.J.; Hawkins, J.S.; Ramirez, C.L.; Batista, L.F.; et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells 2011, 29, 1717–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimatsu, S.; Okahara, J.; Sone, T.; Takeda, Y.; Nakamura, M.; Sasaki, E.; Kishi, N.; Shiozawa, S.; Okano, H. Robust and efficient knock-in in embryonic stem cells and early-stage embryos of the common marmoset using the CRISPR-Cas9 system. Sci. Rep. 2019, 9, 1528. [Google Scholar] [CrossRef] [Green Version]
- Yoshimatsu, S.; Sato, T.; Yamamoto, M.; Sasaki, E.; Nakajima, M.; Nakamura, M.; Shiozawa, S.; Noce, T.; Okano, H. Generation of a male common marmoset embryonic stem cell line DSY127-BV8VT1 carrying double reporters specific for the germ cell linage using the CRISPR-Cas9 and PiggyBac transposase systems. Stem Cell Res. 2020, 44, 101740. [Google Scholar] [CrossRef]
- Kremers, G.J.; Goedhart, J.; van Munster, E.B.; Gadella, T.W., Jr. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 2006, 45, 6570–6580. [Google Scholar] [CrossRef]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.C.; Shao, R.; Pao, A.Y.; Zhang, S.; Hung, M.C.; Su, L.K. Inhibition of cancer cell growth by BRCA2. Cancer Res. 2002, 62, 1311–1314. [Google Scholar]
- Sasaki, E.; Hanazawa, K.; Kurita, R.; Akatsuka, A.; Yoshizaki, T.; Ishii, H.; Tanioka, Y.; Ohnishi, Y.; Suemizu, H.; Sugawara, A.; et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus). Stem Cells 2005, 23, 1304–1313. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Imaizumi, Y.; Okada, Y.; Akamatsu, W.; Koike, M.; Kuzumaki, N.; Hayakawa, H.; Nihira, T.; Kobayashi, T.; Ohyama, M.; Sato, S.; et al. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain 2012, 5, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, T.; Fujimori, K.; Andoh-Noda, T.; Ando, T.; Kuzumaki, N.; Toyoshima, M.; Tada, H.; Imaizumi, K.; Ishikawa, M.; Yamaguchi, R.; et al. Functional Neurons Generated from T Cell-Derived Induced Pluripotent Stem Cells for Neurological Disease Modeling. Stem Cell Rep. 2016, 6, 422–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, A.P.; Bradley, A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 1990, 62, 1073–1085. [Google Scholar] [CrossRef]
- Nishimura, K.; Oki, T.; Kitaura, J.; Kuninaka, S.; Saya, H.; Sakaue-Sawano, A.; Miyawaki, A.; Kitamura, T. APC(CDH1) targets MgcRacGAP for destruction in the late M phase. PLoS ONE 2013, 8, e63001. [Google Scholar] [CrossRef]
- Yaglom, J.A.; McFarland, C.; Mirny, L.; Sherman, M.Y. Oncogene-triggered suppression of DNA repair leads to DNA instability in cancer. Oncotarget 2014, 5, 8367–8378. [Google Scholar] [CrossRef] [Green Version]
- Shiozawa, S.; Nakajima, M.; Okahara, J.; Kuortaki, Y.; Kisa, F.; Yoshimatsu, S.; Nakamura, M.; Koya, I.; Yoshimura, M.; Sasagawa, Y.; et al. Primed to naive-like conversion of the common marmoset embryonic stem cells. Stem Cells Dev. 2020, 29, 761–773. [Google Scholar] [CrossRef]
- Ma, H.; Morey, R.; O’Neil, R.C.; He, Y.; Daughtry, B.; Schultz, M.D.; Hariharan, M.; Nery, J.R.; Castanon, R.; Sabatini, K.; et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 2014, 511, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Torres, R.J.; Puig, J.G. Hypoxanthine-guanine phosophoribosyltransferase (HPRT) deficiency: Lesch-Nyhan syndrome. Orphanet J. Rare Dis. 2007, 2, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, J.D.; Capecchi, N.E.; Capecchi, M.R. Altered enzymes in drug-resistant variants of mammalian tissue culture cells. Proc. Natl. Acad. Sci. USA 1973, 70, 3145–3149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahl, G.M.; Hughes, S.H.; Capecchi, M.R. Immunological characterization of hypoxanthine-guanine phosphoribosyl transferase mutants of mouse L cells: Evidence for mutations at different loci in the HGPRT gene. J. Cell Physiol. 1975, 85, 307–320. [Google Scholar] [CrossRef]
- Thomas, K.R.; Capecchi, M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987, 51, 503–512. [Google Scholar] [CrossRef]
- Meek, S.; Buehr, M.; Sutherland, L.; Thomson, A.; Mullins, J.J.; Smith, A.J.; Burdon, T. Efficient gene targeting by homologous recombination in rat embryonic stem cells. PLoS ONE 2010, 5, e14225. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshimatsu, S.; Nakajima, M.; Qian, E.; Sanosaka, T.; Sato, T.; Okano, H. Homologous Recombination-Enhancing Factors Identified by Comparative Transcriptomic Analyses of Pluripotent Stem Cell of Human and Common Marmoset. Cells 2022, 11, 360. https://doi.org/10.3390/cells11030360
Yoshimatsu S, Nakajima M, Qian E, Sanosaka T, Sato T, Okano H. Homologous Recombination-Enhancing Factors Identified by Comparative Transcriptomic Analyses of Pluripotent Stem Cell of Human and Common Marmoset. Cells. 2022; 11(3):360. https://doi.org/10.3390/cells11030360
Chicago/Turabian StyleYoshimatsu, Sho, Mayutaka Nakajima, Emi Qian, Tsukasa Sanosaka, Tsukika Sato, and Hideyuki Okano. 2022. "Homologous Recombination-Enhancing Factors Identified by Comparative Transcriptomic Analyses of Pluripotent Stem Cell of Human and Common Marmoset" Cells 11, no. 3: 360. https://doi.org/10.3390/cells11030360





