Midbody Proteins Display Distinct Dynamics during Cytokinesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Fluorescence Microscopy
2.3. Western Blot
2.4. Antibodies
2.5. Time-Lapse Imaging
2.6. Computational and Statistical Analyses
3. Results and Discussion
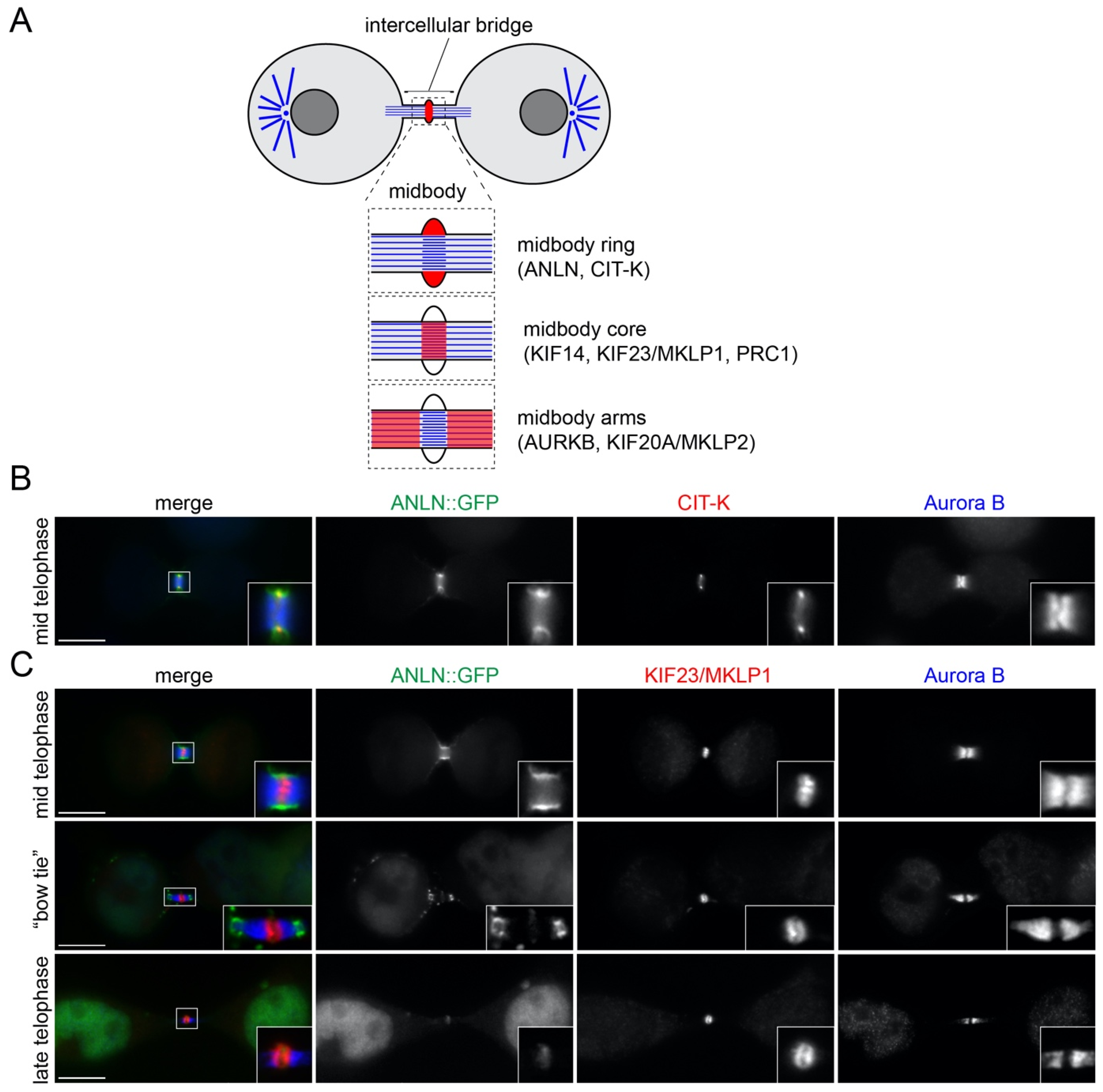
3.1. Midbody Protein Distribution Changes during Midbody Maturation
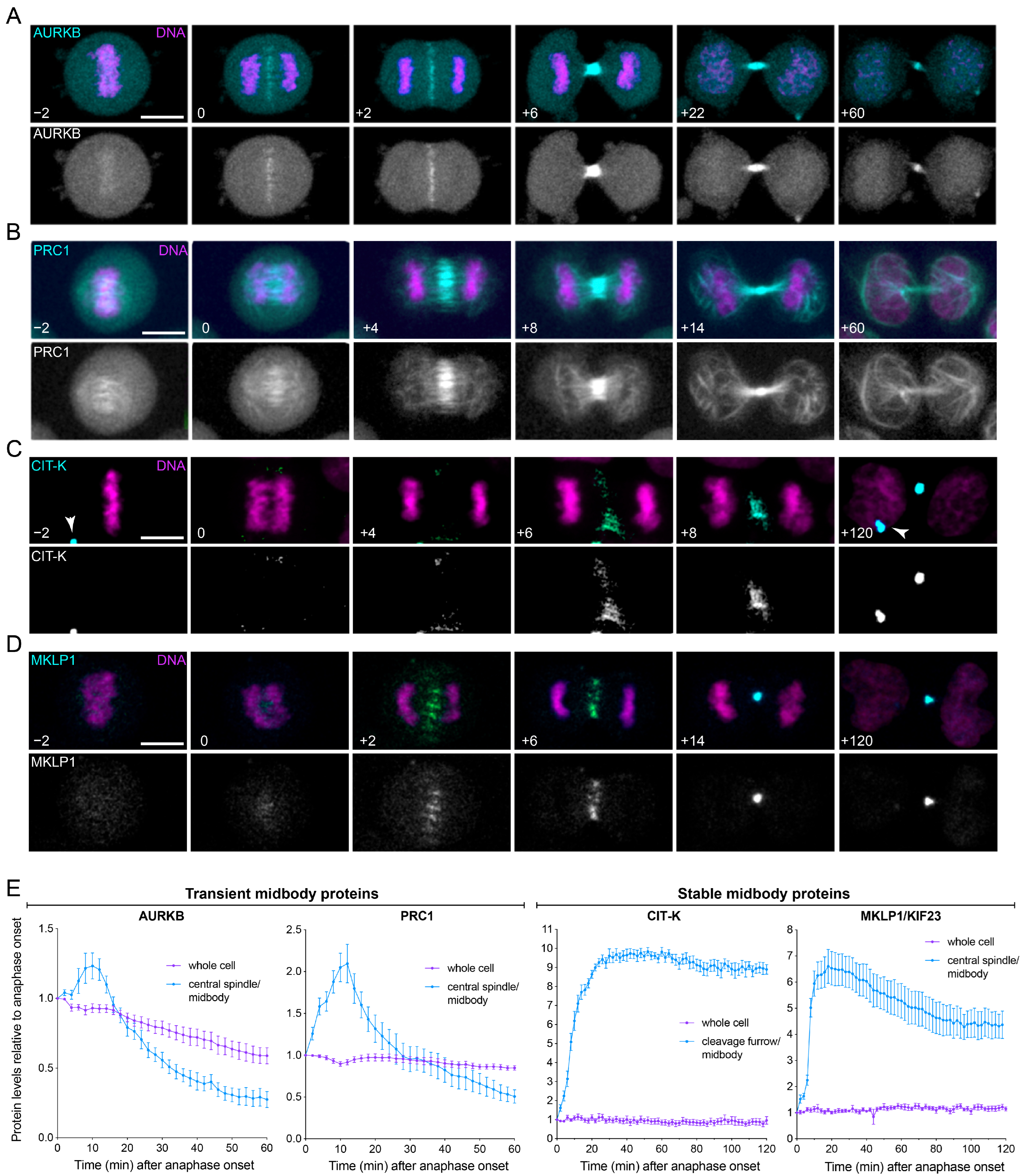
3.2. Midbody Proteins Display Different Expression Profiles during Mitotic Exit and Cytokinesis
3.3. Midbody Proteins Display Distinct Dynamics during Mitotic Exit and Cytokinesis
3.4. Transient and Stable Midbody Proteins Display Distinct Interactions with Ubiquitylation Factors
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cuijpers, S.A.G.; Vertegaal, A.C.O. Guiding Mitotic Progression by Crosstalk between Post-translational Modifications. Trends Biochem. Sci. 2018, 43, 251–268. [Google Scholar] [CrossRef]
- Wieser, S.; Pines, J. The biochemistry of mitosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a015776. [Google Scholar] [CrossRef]
- Lindon, C. Control of mitotic exit and cytokinesis by the APC/C. Biochem. Soc. Trans. 2008, 36, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Holder, J.; Poser, E.; Barr, F.A. Getting out of mitosis: Spatial and temporal control of mitotic exit and cytokinesis by PP1 and PP2A. FEBS Lett. 2019, 593, 2908–2924. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P.; Capalbo, L. New Auroras on the Roles of the Chromosomal Passenger Complex in Cytokinesis: Implications for Cancer Therapies. Front. Oncol. 2015, 5, 221. [Google Scholar] [CrossRef]
- Nasa, I.; Kettenbach, A.N. Coordination of Protein Kinase and Phosphoprotein Phosphatase Activities in Mitosis. Front. Cell Dev. Biol. 2018, 6, 30. [Google Scholar] [CrossRef]
- Archambault, V.; Glover, D.M. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 2009, 10, 265–275. [Google Scholar] [CrossRef]
- Fededa, J.P.; Gerlich, D.W. Molecular control of animal cell cytokinesis. Nat. Cell Biol. 2012, 14, 440–447. [Google Scholar] [CrossRef]
- D’Avino, P.P.; Giansanti, M.G.; Petronczki, M. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 2015, 7, a015834. [Google Scholar] [CrossRef]
- Capalbo, L.; Bassi, Z.I.; Geymonat, M.; Todesca, S.; Copoiu, L.; Enright, A.J.; Callaini, G.; Riparbelli, M.G.; Yu, L.; Choudhary, J.S.; et al. The midbody interactome reveals unexpected roles for PP1 phosphatases in cytokinesis. Nat. Commun. 2019, 10, 4513. [Google Scholar] [CrossRef] [PubMed]
- Skop, A.R.; Liu, H.; Yates, J., 3rd; Meyer, B.J.; Heald, R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 2004, 305, 61–66. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P.; Capalbo, L. Regulation of midbody formation and function by mitotic kinases. Semin. Cell Dev. Biol. 2016, 53, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mierzwa, B.; Gerlich, D.W. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell 2014, 31, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.K.; Coughlin, M.; Mitchison, T.J. Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell 2012, 23, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Bassi, Z.I.; Audusseau, M.; Riparbelli, M.G.; Callaini, G.; D’Avino, P.P. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc. Natl. Acad. Sci. USA 2013, 110, 9782–9787. [Google Scholar] [CrossRef]
- D’Avino, P.P. Citron kinase-renaissance of a neglected mitotic kinase. J. Cell Sci. 2017, 130, 1701–1708. [Google Scholar] [CrossRef]
- McKenzie, C.; Bassi, Z.I.; Debski, J.; Gottardo, M.; Callaini, G.; Dadlez, M.; D’Avino, P.P. Cross-regulation between Aurora B and Citron kinase controls midbody architecture in cytokinesis. Open Biol. 2016, 6, 160019. [Google Scholar] [CrossRef]
- Crowell, E.F.; Gaffuri, A.L.; Gayraud-Morel, B.; Tajbakhsh, S.; Echard, A. Engulfment of the midbody remnant after cytokinesis in mammalian cells. J. Cell Sci. 2014, 127, 3840–3851. [Google Scholar] [CrossRef]
- Peterman, E.; Prekeris, R. The postmitotic midbody: Regulating polarity, stemness, and proliferation. J. Cell Biol. 2019, 218, 3903–3911. [Google Scholar] [CrossRef]
- Siskos, N.; Stylianopoulou, E.; Skavdis, G.; Grigoriou, M.E. Molecular Genetics of Microcephaly Primary Hereditary: An Overview. Brain Sci. 2021, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Hummer, S.; Mayer, T.U. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 2009, 19, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, M.J.; Liu, J.; Lavoie, B.D.; Wilde, A. Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol. 2014, 4, 130190. [Google Scholar] [CrossRef] [PubMed]
- Gai, M.; Camera, P.; Dema, A.; Bianchi, F.; Berto, G.; Scarpa, E.; Germena, G.; Di Cunto, F. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell 2011, 22, 3768–3778. [Google Scholar] [CrossRef] [PubMed]
- Gruneberg, U.; Neef, R.; Li, X.; Chan, E.H.; Chalamalasetty, R.B.; Nigg, E.A.; Barr, F.A. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 2006, 172, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.J.; Suvarna, K.S.; D’Avino, P.P. Synchronization of human retinal pigment ephitilial-1 (RPE-1) cells in mitosis. J. Cell Sci. 2020, 133, jcs247940. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Matsuura, N.; Ubukata, M.; Oikawa, H.; Shima, H.; Kikuchi, K. Tautomycetin is a novel and specific inhibitor of serine/threonine protein phosphatase type 1, PP1. Biochem. Biophys. Res. Commun. 2001, 287, 328–331. [Google Scholar] [CrossRef]
- Ditchfield, C.; Johnson, V.L.; Tighe, A.; Ellston, R.; Haworth, C.; Johnson, T.; Mortlock, A.; Keen, N.; Taylor, S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003, 161, 267–280. [Google Scholar] [CrossRef]
- Zhao, W.M.; Fang, G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J. Biol. Chem. 2005, 280, 33516–33524. [Google Scholar] [CrossRef]
- Paccosi, E.; Costanzo, F.; Costantino, M.; Balzerano, A.; Monteonofrio, L.; Soddu, S.; Prantera, G.; Brancorsini, S.; Egly, J.M.; Proietti-De-Santis, L. The Cockayne syndrome group A and B proteins are part of a ubiquitin-proteasome degradation complex regulating cell division. Proc. Natl. Acad. Sci. USA 2020, 117, 30498–30508. [Google Scholar] [CrossRef] [PubMed]
- Glotzer, M.; Murray, A.W.; Kirschner, M.W. Cyclin is degraded by the ubiquitin pathway. Nature 1991, 349, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.; Pines, J.; Lindon, C. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr. Biol. 2008, 18, 1649–1658. [Google Scholar] [CrossRef]
- Watanabe, S.; De Zan, T.; Ishizaki, T.; Narumiya, S. Citron kinase mediates transition from constriction to abscission through its coiled-coil domain. J. Cell Sci. 2013, 126, 1773–1784. [Google Scholar] [CrossRef]
- Galarreta, A.; Valledor, P.; Ubieto-Capella, P.; Lafarga, V.; Zarzuela, E.; Munoz, J.; Malumbres, M.; Lecona, E.; Fernandez-Capetillo, O. USP7 limits CDK1 activity throughout the cell cycle. EMBO J. 2021, 40, e99692. [Google Scholar] [CrossRef] [PubMed]
- Giovinazzi, S.; Morozov, V.M.; Summers, M.K.; Reinhold, W.C.; Ishov, A.M. USP7 and Daxx regulate mitosis progression and taxane sensitivity by affecting stability of Aurora-A kinase. Cell Death Differ. 2013, 20, 721–731. [Google Scholar] [CrossRef]
- Giovinazzi, S.; Sirleto, P.; Aksenova, V.; Morozov, V.M.; Zori, R.; Reinhold, W.C.; Ishov, A.M. Usp7 protects genomic stability by regulating Bub3. Oncotarget 2014, 5, 3728–3742. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, Y.; Gao, Y.; Yuan, B.; Qi, X.; Fu, Y.; Zhu, Q.; Cao, T.; Zhang, S.; Yin, L.; et al. USP7 is a novel Deubiquitinase sustaining PLK1 protein stability and regulating chromosome alignment in mitosis. J. Exp. Clin. Cancer Res. 2019, 38, 468. [Google Scholar] [CrossRef]
- Skowyra, A.; Allan, L.A.; Saurin, A.T.; Clarke, P.R. USP9X Limits Mitotic Checkpoint Complex Turnover to Strengthen the Spindle Assembly Checkpoint and Guard against Chromosomal Instability. Cell Rep. 2018, 23, 852–865. [Google Scholar] [CrossRef]
- Vong, Q.P.; Cao, K.; Li, H.Y.; Iglesias, P.A.; Zheng, Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 2005, 310, 1499–1504. [Google Scholar] [CrossRef]

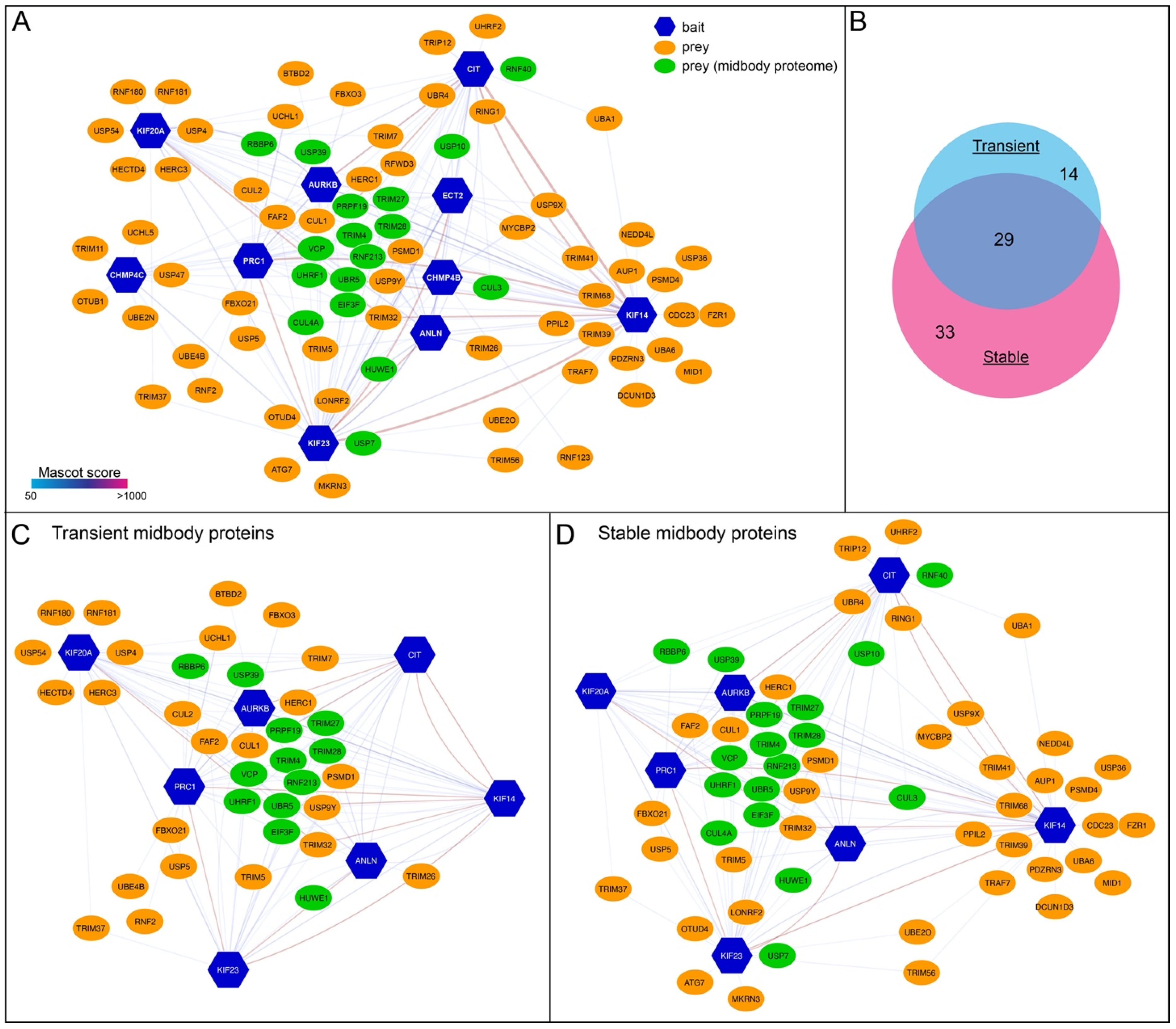
| Group | Gene Name | Protein Name | Baits (MASCOT Score) | Midbody Proteome |
|---|---|---|---|---|
| Common | VCP | Transitional endoplasmic reticulum ATPase (TER ATPase) | ANLN (84); AURKB (174); KIF14 (76); PRC1 (158) | Yes |
| USP5 | Ubiquitin carboxyl-terminal hydrolase 5 | MKLP1 (41); PRC1 (83) | No | |
| EIF3F | Eukaryotic translation initiation factor 3 subunit F | ANLN (61); CIT-K (79); KIF14 (59); MKLP1 (73); PRC1 (136) | Yes | |
| USP9Y | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y | KIF14 (42); KIF20A (36); MKLP1 (68) | No | |
| USP39 | U4/U6.U5 tri-snRNP-associated protein 2 | AURKB (36); CIT-K (300); PRC1 (156) | Yes | |
| Transient-specific | UCHL1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | KIF20A (30); PRC1 (392) | No |
| USP54 | Inactive ubiquitin carboxyl-terminal hydrolase 54 | KIF20A (104) | No | |
| USP4 | Ubiquitin carboxyl-terminal hydrolase 4 | KIF20A (40) | No | |
| Stable-specific | OTUD4 | OTU domain-containing protein 4 | MKLP1 (38) | No |
| USP7 | Ubiquitin carboxyl-terminal hydrolase 7 | MKLP1 (51) | Yes | |
| USP10 | Ubiquitin carboxyl-terminal hydrolase 10 | CIT-K (59); KIF14 (126) | Yes | |
| USP36 | Ubiquitin carboxyl-terminal hydrolase 36 | KIF14 (139) | No | |
| USP9X | Probable ubiquitin carboxyl-terminal hydrolase FAF-X | KIF14 (50) | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halcrow, E.F.J.; Mazza, R.; Diversi, A.; Enright, A.; D’Avino, P.P. Midbody Proteins Display Distinct Dynamics during Cytokinesis. Cells 2022, 11, 3337. https://doi.org/10.3390/cells11213337
Halcrow EFJ, Mazza R, Diversi A, Enright A, D’Avino PP. Midbody Proteins Display Distinct Dynamics during Cytokinesis. Cells. 2022; 11(21):3337. https://doi.org/10.3390/cells11213337
Chicago/Turabian StyleHalcrow, Ella F. J., Riccardo Mazza, Anna Diversi, Anton Enright, and Pier Paolo D’Avino. 2022. "Midbody Proteins Display Distinct Dynamics during Cytokinesis" Cells 11, no. 21: 3337. https://doi.org/10.3390/cells11213337
APA StyleHalcrow, E. F. J., Mazza, R., Diversi, A., Enright, A., & D’Avino, P. P. (2022). Midbody Proteins Display Distinct Dynamics during Cytokinesis. Cells, 11(21), 3337. https://doi.org/10.3390/cells11213337






