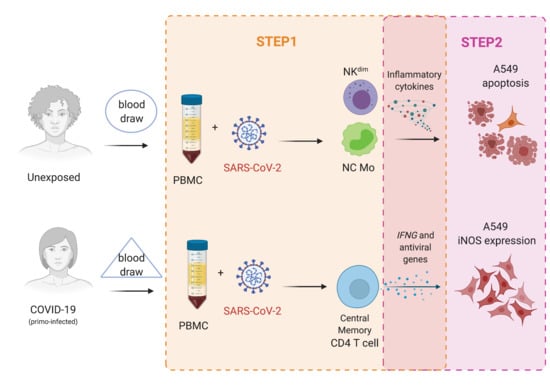
Two-Step In Vitro Model to Evaluate the Cellular Immune Response to SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Samples
Ethical Statement
2.3. Laboratory Assays
2.3.1. Viral Detection by RT-qPCR
2.3.2. Step1: In Vitro Antigen-Specific Cellular Immune Response
2.3.3. Cytokines/Chemokines Gene Expression
2.3.4. Step2: Human Alveolar Lung Epithelial Cells (A549) Cultivation with Supernatant from PBMC Cultured with SARS-CoV-2
2.4. Statistical Analysis
3. Results
3.1. SARS-CoV-2 Replication on Mononuclear Cells Subsets
3.2. Step1: SARS-CoV-2 Effects on the Functionality of PBMC Phenotypes and Cytokine Gene Expression
3.2.1. Natural Killer Cells
3.2.2. Monocytes
3.2.3. CD4+ T and CD8+ T Lymphocytes
3.2.4. Memory T Lymphocytes
3.2.5. Expression of SARS-CoV-2 Response Genes
3.3. Step2: Effects of Supernatant from PBMC Cell Cultivation in Lung Alveolar Epithelial Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Yang, B.; Li, Q.; Wen, L.; Zhang, R. Clinical Features of 69 Cases with Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020, 71, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef]
- Frederiksen, L.S.F.; Zhang, Y.; Foged, C.; Thakur, A. The Long Road toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol. 2020, 11, 1817. [Google Scholar] [CrossRef]
- Fontanet, A.; Cauchemez, S. COVID-19 herd immunity: Where are we? Nat. Rev. Immunol. 2020, 20, 583–584. [Google Scholar] [CrossRef]
- Chung, J.Y.; Thone, M.N.; Kwon, Y.J. COVID-19 vaccines: The status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021, 170, 1–25. [Google Scholar] [CrossRef]
- Giordano, G.; Colaneri, M.; Di Filippo, A.; Blanchini, F.; Bolzern, P.; De Nicolao, G.; Sacchi, P.; Colaneri, P.; Bruno, R. Modeling vaccination rollouts, SARS-CoV-2 variants and the requirement for non-pharmaceutical interventions in Italy. Nat. Med. 2021, 27, 993–998. [Google Scholar] [CrossRef]
- Grech, V.; Souness, J.; Agius, S. Mass population vaccination for COVID-19 in Malta. J. Vis. Commun. Med. 2021, 1–7. [Google Scholar] [CrossRef]
- Chibber, P.; Haq, S.A.; Ahmed, I.; Andrabi, N.I.; Singh, G. Advances in the possible treatment of COVID-19: A review. Eur. J. Pharm. 2020, 883, 173372. [Google Scholar] [CrossRef]
- Newton, A.H.; Cardani, A.; Braciale, T.J. The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin. Immunopathol. 2016, 38, 471–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Julian, B.; Lucie, L.; Marco, F.; Daniel, W.; Philipp, G.; Florian, K.; Stefan, H.; Manuela, D.; Beate, K.; Florent, F.; et al. Presence of SARS-CoV-2-reactive T cells in COVID-19 patients and healthy donors. medRxiv 2020, 20061440. [Google Scholar] [CrossRef]
- Chen, Z.; Wherry, E.J. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020, 20, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Pontelli, M.C.; Castro, I.A.; Martins, R.B.; Veras, F.P.; La Serra, L.; Nascimento, D.C.; Cardoso, R.S.; Rosales, R.; Lima, T.M.; Souza, J.P.; et al. Infection of human lymphomononuclear cells by SARS-CoV-2. bioRxiv 2020, 225912. [Google Scholar] [CrossRef]
- Van Elslande, J.; Vermeersch, P.; Vandervoort, K.; Wawina-Bokalanga, T.; Vanmechelen, B.; Wollants, E.; Laenen, L.; André, E.; Van Ranst, M.; Lagrou, K.; et al. Symptomatic Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection by a Phylogenetically Distinct Strain. Clin. Infect. Dis. 2021, 73, 354–356. [Google Scholar] [CrossRef]
- To, K.K.-W.; Hung, I.F.-N.; Ip, J.D.; Chu, A.W.-H.; Chan, W.-M.; Tam, A.R.; Fong, C.H.-Y.; Yuan, S.; Tsoi, H.-W.; Ng, A.C.-K.; et al. Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. 2020, ciaa1275. [Google Scholar] [CrossRef] [PubMed]
- Tillett, R.L.; Sevinsky, J.R.; Hartley, P.D.; Kerwin, H.; Crawford, N.; Gorzalski, A.; Laverdure, C.; Verma, S.C.; Rossetto, C.C.; Jackson, D.; et al. Genomic evidence for reinfection with SARS-CoV-2: A case study. Lancet Infect. Dis. 2021, 21, 52–58. [Google Scholar] [CrossRef]
- Ota, M. Will we see protection or reinfection in COVID-19? Nat. Rev. Immunol. 2020, 20, 351. [Google Scholar] [CrossRef]
- Gousseff, M.; Penot, P.; Gallay, L.; Batisse, D.; Benech, N.; Bouiller, K.; Collarino, R.; Conrad, A.; Slama, D.; Joseph, C.; et al. Clinical recurrences of COVID-19 symptoms after recovery: Viral relapse, reinfection or inflammatory rebound? J. Infect. 2020, 81, 816–846. [Google Scholar] [CrossRef]
- Roy, S. COVID-19 Reinfection: Myth or Truth? SN Compr. Clin. Med. 2020, 2, 710–713. [Google Scholar] [CrossRef]
- Jabbari, P.; Rezaei, N. With Risk of Reinfection, Is COVID-19 Here to Stay? Disaster Med. Public Health Prep. 2020, 14, e33. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Thevarajan, I.; Nguyen, T.H.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses underpinning viral clearance and patient recovery in a non-severe case of COVID-19. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos, A.D.R.; Motta, F.C.; Caetano, B.C.; Ogrzewalska, M.; Garcia, C.C.; Lopes, J.C.O.; Miranda, M.; Livorati, M.T.F.P.; Abreu, A.; Brown, D.; et al. Identification of SARS-CoV-2 and additional respiratory pathogens cases under the investigation of COVID-19 initial phase in a Brazilian reference laboratory. Memórias Inst. Oswaldo Cruz. 2020, 115, 200232. [Google Scholar] [CrossRef]
- Paola, C.R.; Edson, D.; Tiago, G.; Daiana, M.; Fernando, d.C.M.; Luciana, R.A.; Anna, C.D.d.P.; Maria, O.; Braulia, C.; Mirleide, C.S.; et al. Genomic surveillance of SARS-CoV-2 reveals community transmission of a major lineage during the early pandemic phase in Brazil. Evol. Biol. 2020. [Google Scholar] [CrossRef]
- Son, K.-N.; Liang, Z.; Lipton, H.L. Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses. J. Virol. 2015, 89, 9383–9392. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, L.T.A.; Robottom-Ferreira, A.B.; Ribeiro-Alves, M.; Toledo-Pinto, T.G.; Brito, T.R.; Rosa, P.S.; Sandoval, F.G.; Jardim, M.R.; Antunes, S.G.; Shannon, E.J.; et al. Gene Expression Profiling Specifies Chemokine, Mitochondrial and Lipid Metabolism Signatures in Leprosy. PLoS ONE 2013, 8, e64748. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Gil Melgaço, J.; E Cunha, D.B.; Azamor, T.; Da Silva, A.M.V.; Tubarão, L.N.; Gonçalves, R.B.; Monteiro, R.Q.; Missailidis, S.; Neves, P.C.D.C.; Bom, A.P.D.A. Cellular and Molecular Immunology Approaches for the Development of Immunotherapies against the New Coronavirus (SARS-CoV-2): Challenges to Near-Future Breakthroughs. J. Immunol. Res. 2020, 2020, 1–21. [Google Scholar] [CrossRef]
- Peng, L.; Liu, J.; Xu, W.; Luo, Q.; Chen, D.; Lei, Z.; Huang, Z.; Li, X.; Deng, K.; Lin, B.; et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020, 92, 1676–1680. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.; Liu, H.; Li, W.; Lin, F.; Jiang, L.; Li, X.; Xu, P.; Zhang, L.; Zhao, L.; et al. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 2020, 73, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.-F.; Gong, H.-R.; Lee, A.C.-Y.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020, 30, 928–931. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Xu, L.; Xie, X.; Yan, H.; Xie, B.; Xu, W.; Liu, X.; Kang, G.; Jiang, W.; Yuan, J. Pulmonary pathology of early-phase COVID-19 pneumonia in a patient with a benign lung lesion. Histopathology 2020, 77, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Brownlie, D.; et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020, 5, eabd6832. [Google Scholar] [CrossRef]
- Gatti, A.; Radrizzani, D.; Viganò, P.; Mazzone, A.; Brando, B. Decrease of Non-Classical and Intermediate Monocyte Subsets in Severe Acute SARS-CoV -2 Infection. Cytom. Part A 2020, 97, 887–890. [Google Scholar] [CrossRef]
- Natalia, F.R.; Carolina, Q.S.; Carlyle, R.L.; Franklin, S.S.; André, C.F.; Mayara, M.; Caroline, S.F.; Vinicius, C.S.; Suelen, S.G.D.; Jairo, R.T.; et al. Atazanavir inhibits SARS-CoV-2 replication and pro-inflammatory cytokine production. bioRxiv 2020, 020925. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Qi, T.; Liu, L.; Ling, Y.; Qian, Z.; Li, T.; Li, F.; Xu, Q.; Zhang, Y.; Xu, S.; et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020, 80, e1–e6. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.-Y.; Zhang, M.; Yang, C.-X.; Zhang, N.; Wang, X.-C.; Yang, X.-P.; Dong, X.-Q.; Zheng, Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef]
- López-Collazo, E.; Avendaño-Ortiz, J.; Martín-Quirós, A.; Aguirre, L.A. Immune Response and COVID-19: A mirror image of Sepsis. Int. J. Biol. Sci. 2020, 16, 2479–2489. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, L.; Lin, C. T cell response in patients with COVID-19. Blood Sci. 2020, 2, 76–78. [Google Scholar] [CrossRef]
- Ducloyer, M.; Gaborit, B.; Toquet, C.; Castain, L.; Bal, A.; Arrigoni, P.P.; LeComte, R.; Clement, R.; Sagan, C. Complete post-mortem data in a fatal case of COVID-19: Clinical, radiological and pathological correlations. Int. J. Leg. Med. 2020, 134, 2209–2214. [Google Scholar] [CrossRef]
- Nienhold, R.; Ciani, Y.; Koelzer, V.H.; Tzankov, A.; Haslbauer, J.D.; Menter, T.; Schwab, N.; Henkel, M.; Frank, A.; Zsikla, V.; et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Thieme, C.J.; Anft, M.; Paniskaki, K.; Blazquez-Navarro, A.; Doevelaar, A.; Seibert, F.S.; Hoelzer, B.; Konik, M.J.; Berger, M.M.; Brenner, T.; et al. Robust T Cell Response Toward Spike, Membrane, and Nucleocapsid SARS-CoV-2 Proteins Is Not Associated with Recovery in Critical COVID-19 Patients. Cell Rep. Med. 2020, 1, 100092. [Google Scholar] [CrossRef]
- Zuo, J.; Dowell, A.C.; Pearce, H.; Verma, K.; Long, H.M.; Begum, J.; Aiano, F.; Amin-Chowdhury, Z.; Hoschler, K.; Brooks, T.; et al. Robust SARS-CoV-2-specific T-cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2020, 22, 620–626. [Google Scholar] [CrossRef]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Macary, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Gil Melgaço, J.; Azamor, T.; Bom, A.P.D.A. Protective immunity after COVID-19 has been questioned: What can we do without SARS-CoV-2-IgG detection? Cell. Immunol. 2020, 353, 104114. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, N.; Kuo, H.-H.; Boucau, J.; Farmer, J.R.; Allard-Chamard, H.; Mahajan, V.S.; Piechocka-Trocha, A.; Lefteri, K.; Osborn, M.; Bals, J.; et al. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell 2020, 183, 143–157.e13. [Google Scholar] [CrossRef]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; Sette, A.; et al. Single-Cell Transcriptomic Analysis of SARS-CoV-2 Reactive CD4 + T Cells. Soc. Sci. Res. Netw. 2020, 7, 3641939. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020, 7, 998–1002. [Google Scholar] [CrossRef] [Green Version]
- Pia, L. SARS-CoV-2-reactive T cells in patients and healthy donors. Nat. Rev. Immunol. 2020, 20, 353. [Google Scholar] [CrossRef]
- Oestreich, K.J.; Weinmann, A.S. Transcriptional mechanisms that regulate T helper 1 cell differentiation. Curr. Opin. Immunol. 2012, 24, 191–195. [Google Scholar] [CrossRef] [Green Version]
- De Araújo-Souza, P.S.; Hanschke, S.C.H.; Viola, J. Epigenetic Control of Interferon-Gamma Expression in CD8 T Cells. J. Immunol. Res. 2015, 2015, 849573. [Google Scholar] [CrossRef] [Green Version]
- Pulko, V.; Davies, J.S.; Martinez, C.; Lanteri, M.C.; Busch, M.C.L.M.P.; Diamond, M.S.; Knox, K.; Bush, E.C.; Sims, P.; Sinari, S.; et al. Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat. Immunol. 2016, 17, 966–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, S.N.; Gebhardt, T.; Carbone, F.R.; Heath, W. Memory T Cell Subsets, Migration Patterns, and Tissue Residence. Annu. Rev. Immunol. 2013, 31, 137–161. [Google Scholar] [CrossRef] [PubMed]
- Raeber, M.E.; Zurbuchen, Y.; Impellizzieri, D.; Boyman, O. The role of cytokines in T-cell memory in health and disease. Immunol. Rev. 2018, 283, 176–193. [Google Scholar] [CrossRef]
- Stephens, D.S.; McElrath, M.J. COVID-19 and the Path to Immunity. JAMA 2020, 324, 1279. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Wang, C.; Jin, S.W.; Labrecque, M.; Beischlag, T.V.; Brockman, M.A.; Choy, J.C. Expression of human inducible nitric oxide synthase in response to cytokines is regulated by hypoxia-inducible factor-1. Free Radic. Biol. Med. 2019, 130, 278–287. [Google Scholar] [CrossRef]
- Kaplan, A.; Ciftci, G.A.; Kutlu, M. The apoptotic and genomic studies on A549 cell line induced by silver nitrate. Tumor Biol. 2017, 39, 101042831769503. [Google Scholar] [CrossRef] [Green Version]
- Chu, S.C.; Marks-Konczalik, J.; Wu, H.-P.; Banks, T.C.; Moss, J. Analysis of the Cytokine-Stimulated Human Inducible Nitric Oxide Synthase (iNOS) Gene: Characterization of Differences between Human and Mouse iNOS Promoters. Biochem. Biophys. Res. Commun. 1998, 248, 871–878. [Google Scholar] [CrossRef]
- Zeng, M.; Huang, C.; Zheng, H.; Chen, Q.; He, W.; Deng, Y. Effects of Ghrelin on iNOS-Derived NO Promoted LPS-Induced Pulmonary Alveolar Epithelial A549 Cells Apoptosis. Cell. Physiol. Biochem. 2018, 49, 1840–1855. [Google Scholar] [CrossRef]
- Korhonen, R.; Linker, K.; Pautz, A.; Förstermann, U.; Moilanen, E.; Kleinert, H. Post-Transcriptional Regulation of Human Inducible Nitric-Oxide Synthase Expression by the Jun N-terminal Kinase. Mol. Pharm. 2007, 71, 1427–1434. [Google Scholar] [CrossRef] [Green Version]
- Stefano, G.B.; Esch, T.; Kream, R.M. Potential Immunoregulatory and Antiviral/SARS-CoV-2 Activities of Nitric Oxide. Med. Sci. Monit. 2020, 26, e925679-1–e925679-3. [Google Scholar] [CrossRef] [PubMed]
- Akaberi, D.; Krambrich, J.; Ling, J.; Luni, C.; Hedenstierna, G.; Järhult, J.D.; Lennerstrand, J.; Lundkvist, Å. Mitigation of the replication of SARS-CoV-2 by nitric oxide in vitro. Redox Biol. 2020, 37, 101734. [Google Scholar] [CrossRef] [PubMed]
- Lange-Consiglio, A.; Gusmara, C.; Manfredi, E.; Idda, A.; Soggiu, A.; Greco, V.; Bonizzi, L.; Cremonesi, F.; Zecconi, A. Antimicrobial Effects of Conditioned Medium From Amniotic Progenitor Cells in vitro and in vivo: Toward Tissue Regenerative Therapies for Bovine Mastitis. Front. Vet. Sci. 2019, 6, 443. [Google Scholar] [CrossRef] [PubMed]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Vendramini, P.H.; Valença, A.G.; Antunes, A.S.; Brandão-Teles, C.; da Silva Zuccoli, G.; Reis-de-Oliveira, G.; Silva-Costa, L.C.; et al. SARS-CoV-2 infects brain astrocytes of COVID-19 patients and impairs neuronal viability. medRxiv 2020. [Google Scholar] [CrossRef]







| ID | Gender | RT-PCR SARS-CoV-2 | Serology IgM/IgG | Signals & Symptoms | Age | Spike Assay (BSL2) | Virus Assay (BSL3) |
|---|---|---|---|---|---|---|---|
| 1 | F | Negative | Negative | No | 45 | Yes | No |
| 2 | M | Positive | Positive | Yes | 44 | Yes | Yes |
| 3 | F | Positive | Negative | Yes | 28 | Yes | No |
| 4 | F | Positive | Negative | Yes | 38 | Yes | No |
| 5 | F | Positive | Negative | Yes | 37 | Yes | No |
| 6 | M | Positive | Negative | Yes | 29 | Yes | No |
| 7 | M | Positive | Positive | Yes | 28 | Yes | No |
| 8 | F | Negative | Negative | No | 32 | Yes | No |
| 9 | F | Positive | Positive | Yes | 32 | Yes | Yes |
| 10 | F | Negative | Negative | No | 29 | Yes | No |
| 11 | M | Negative | Negative | No | 31 | Yes | No |
| 12 | M | Negative | Negative | No | 45 | Yes | Yes |
| 13 | F | Negative | Negative | No | 42 | Yes | Yes |
| 14 | F | Negative | Negative | No | 31 | Yes | Yes |
| 15 | F | Negative | Negative | No | 52 | Yes | No |
| 16 | F | Negative | Negative | No | 39 | Yes | Yes |
| 17 | M | Negative | Negative | No | 26 | Yes | No |
| 18 | M | Negative | Negative | No | 24 | Yes | No |
| 19 | F | Negative | Negative | No | 28 | Yes | Yes |
| 20 | F | Negative | Negative | No | 30 | Yes | No |
| 21 | F | Negative | Negative | No | 36 | Yes | Yes |
| 22 | M | Negative | Negative | No | 38 | Yes | Yes |
| 23 | M | Negative | Negative | No | 38 | Yes | Yes |
| 24 | M | Negative | Negative | No | 31 | Yes | Yes |
| 25 | M | Negative | Negative | No | 29 | Yes | Yes |
| 26 | F | Positive | Positive | Yes | 34 | Yes | Yes |
| 27 | M | Positive | Positive | Yes | 56 | Yes | Yes |
| 28 | F | Positive | Positive | Yes | 58 | Yes | Yes |
| 29 | M | Positive | Positive | Yes | 34 | Yes | Yes |
| Unexposed (n = 18) | COVID-19 (n = 11) | ||
|---|---|---|---|
| RT-qPCR SARS-CoV-2 | Positive | __ | 11 (100%) |
| Negative | 18 (100%) | __ | |
| Serology IgM/IgG | Positive | __ | 7 (63.6%) |
| Negative | 18 (100%) | 4 (36.4%) | |
| Signals and symptoms | Yes | __ | 11 (100%) |
| No | 18 (100%) | __ | |
| Demographic | Gender (F; M) | 10; 8 | 6; 5 |
| Age (mean ± SD) | 35 ± 8 | 38 ± 11 |
| Unexposed (Mean ± SE) | COVID-19 (Mean ± SE) | p Value | |
|---|---|---|---|
| T cells | 36.75 ± 4.03 | 30.7 ± 3.00 | 0.48 |
| B cells | 15.25 ± 4.75 | 8.48 ± 3.38 | 0.22 |
| Monocytes | 2.73 ± 1.68 | 3.31 ± 1.55 | 0.25 |
| Natural killer | 1.61 ± 0.45 | 1.34 ± 0.27 | 0.11 |
| Infected | Noninfected | p Value Infected vs. Noninfected | |
|---|---|---|---|
| Mock A549 | 56.34 ± 3.34 | 1.47 ± 0.32 | 0.018 |
| NC (Unexposed) | 57.82 ± 1.25 | 1.48 ± 0.43 | 0.019 |
| NC (COVID-19) | 53.71 ± 8.32 | 1.40 ± 0.33 | 0.024 |
| Virus (Unexposed) | 54.32 ± 6.43 | 42.2 ± 7.31 | 0.221 |
| Virus (COVID-19) | 55.61 ± 7.82 | 42.3 ± 9.60 | 0.250 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melgaço, J.G.; Azamor, T.; Silva, A.M.V.; Linhares, J.H.R.; dos Santos, T.P.; Mendes, Y.S.; de Lima, S.M.B.; Fernandes, C.B.; da Silva, J.; de Souza, A.F.; et al. Two-Step In Vitro Model to Evaluate the Cellular Immune Response to SARS-CoV-2. Cells 2021, 10, 2206. https://doi.org/10.3390/cells10092206
Melgaço JG, Azamor T, Silva AMV, Linhares JHR, dos Santos TP, Mendes YS, de Lima SMB, Fernandes CB, da Silva J, de Souza AF, et al. Two-Step In Vitro Model to Evaluate the Cellular Immune Response to SARS-CoV-2. Cells. 2021; 10(9):2206. https://doi.org/10.3390/cells10092206
Chicago/Turabian StyleMelgaço, Juliana G., Tamiris Azamor, Andréa M. V. Silva, José Henrique R. Linhares, Tiago P. dos Santos, Ygara S. Mendes, Sheila M. B. de Lima, Camilla Bayma Fernandes, Jane da Silva, Alessandro F. de Souza, and et al. 2021. "Two-Step In Vitro Model to Evaluate the Cellular Immune Response to SARS-CoV-2" Cells 10, no. 9: 2206. https://doi.org/10.3390/cells10092206