Dissecting the RecA-(In)dependent Response to Mitomycin C in Mycobacterium tuberculosis Using Transcriptional Profiling and Proteomics Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains Cultures
2.2. Recombinant M. tuberculosis RecA Expression, Purification, and Production of Anti-RecA Rabbit Polyvalent Serum
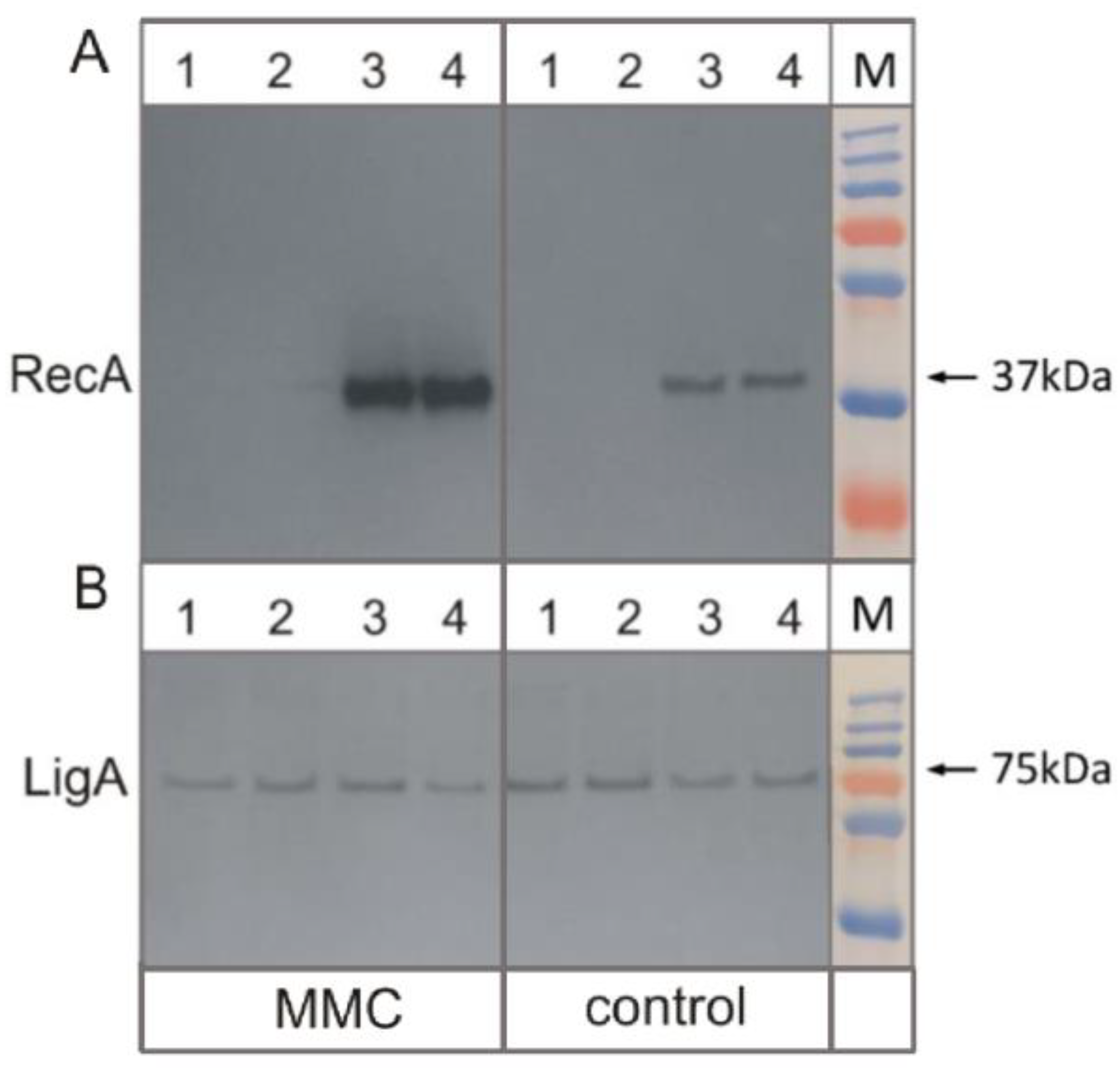
2.3. Total Protein Isolation and Western Blotting
2.4. Survival of M. tuberculosis Strains Exposed to UV Light or in the Presence of Mitomycin C (MMC)
2.5. RNA Isolation and Sequencing
2.6. Transcriptional and Proteomics Data Analysis
2.7. Evolutionary Pressure Analysis
3. Results
3.1. DNA Repair Genes under Evolutionary Pressure
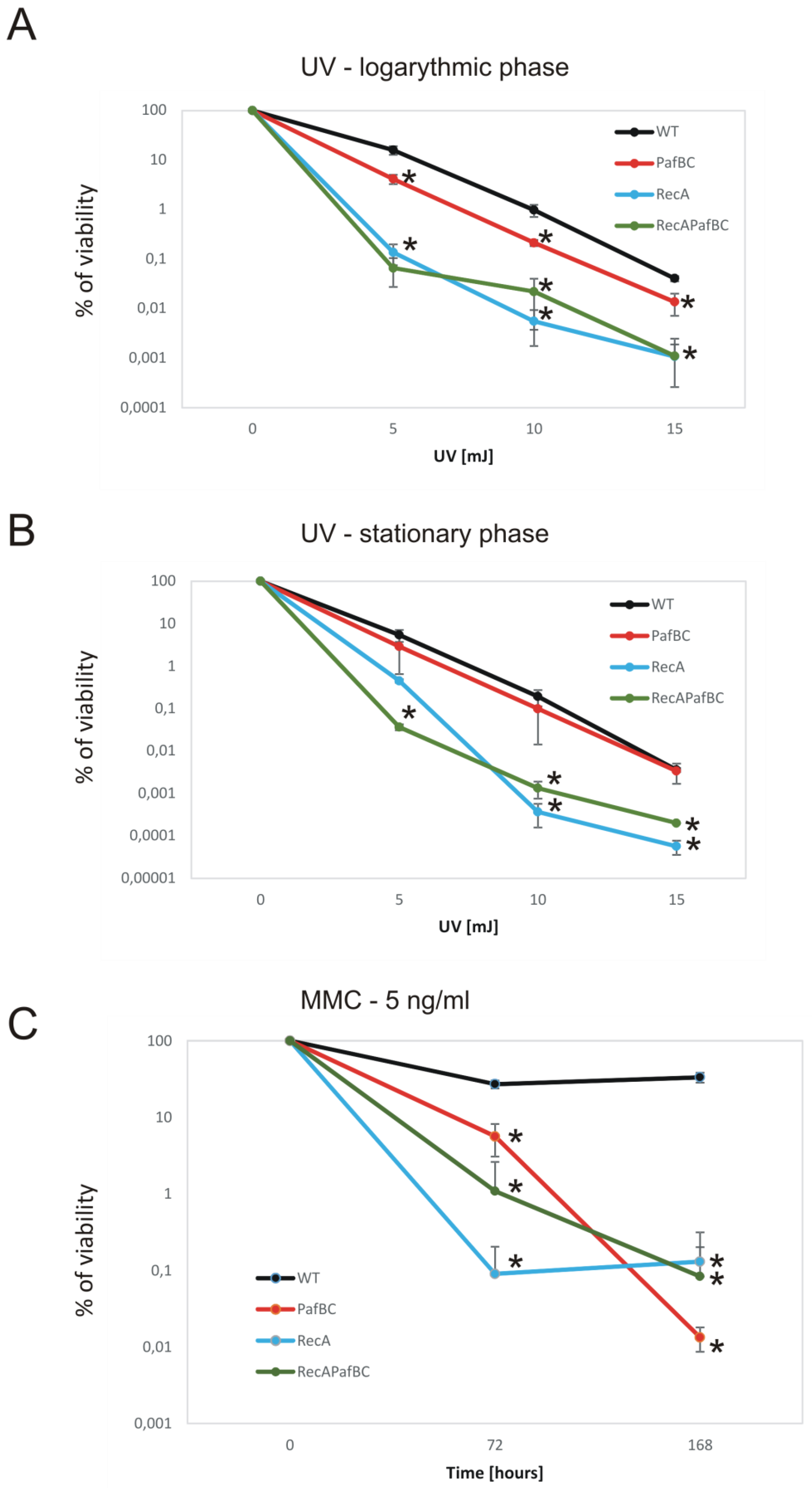
3.2. Deficiency of RecA and/or PafBC Leads to the Sensitization of Tubercle Bacilli to DNA Damage
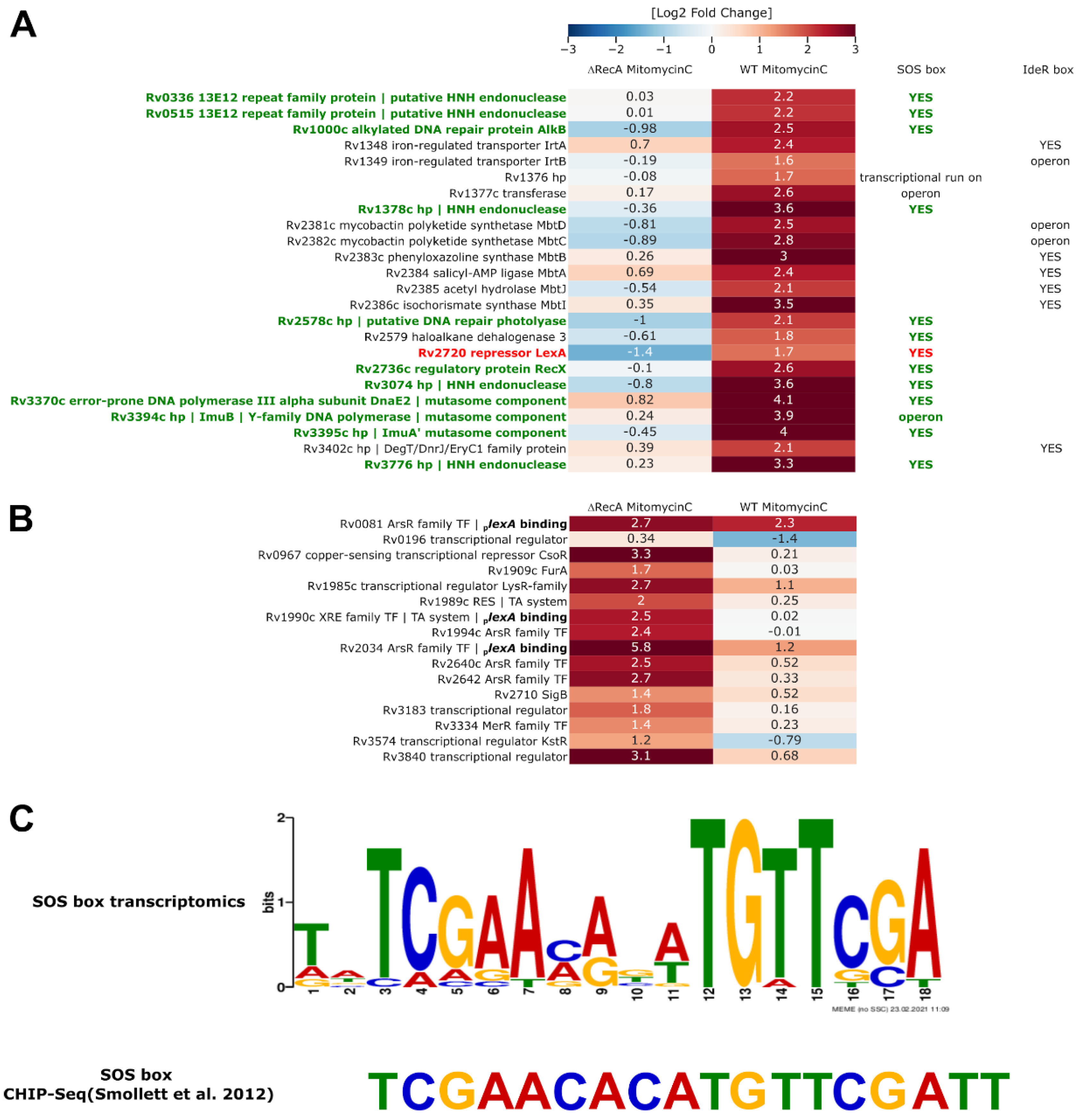
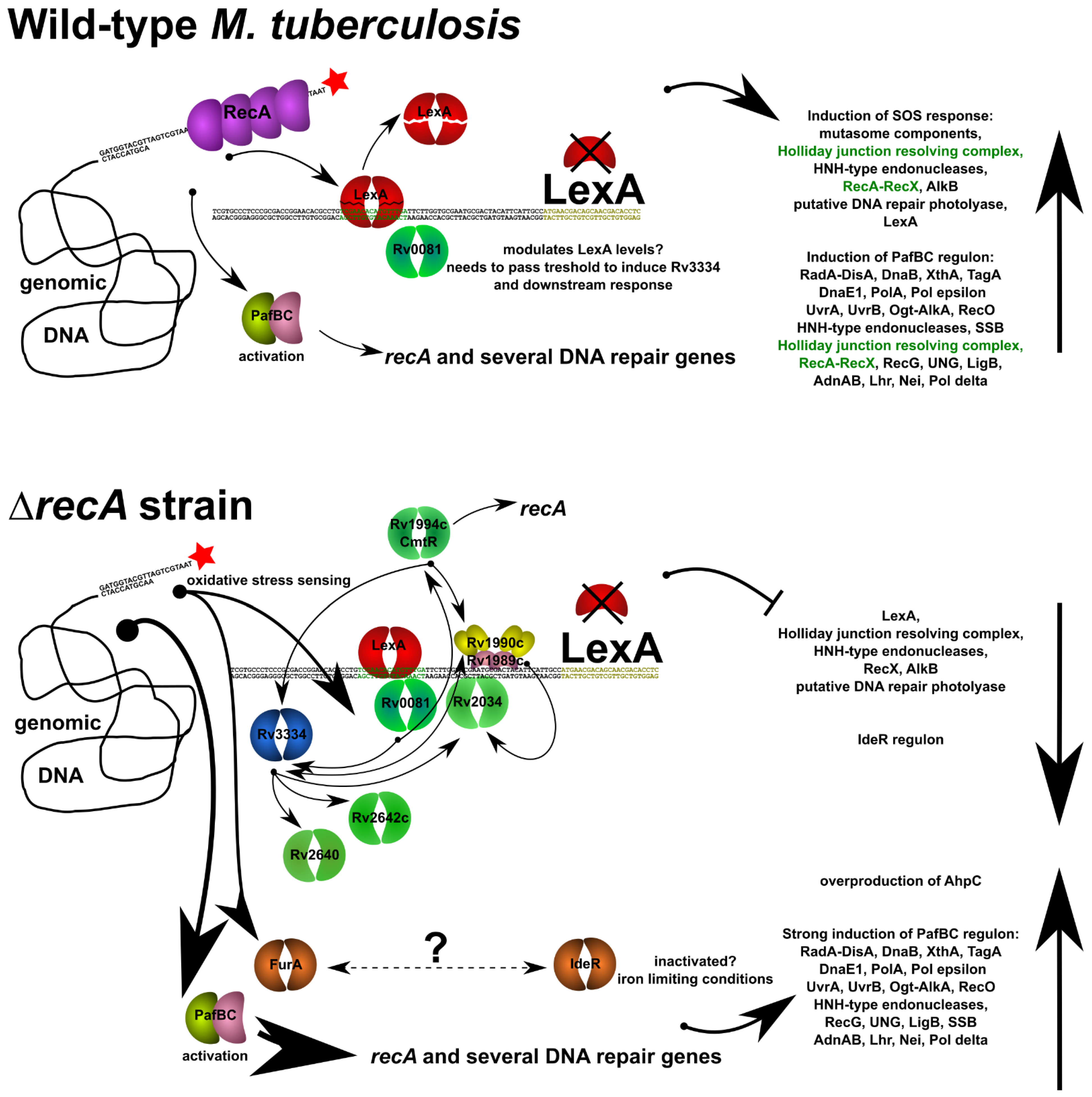
3.3. Removal of RecA Recombinase Leads to Transcriptional Repression of LexA in Response to the DNA Damage-Inducing Agent Mitomycin C
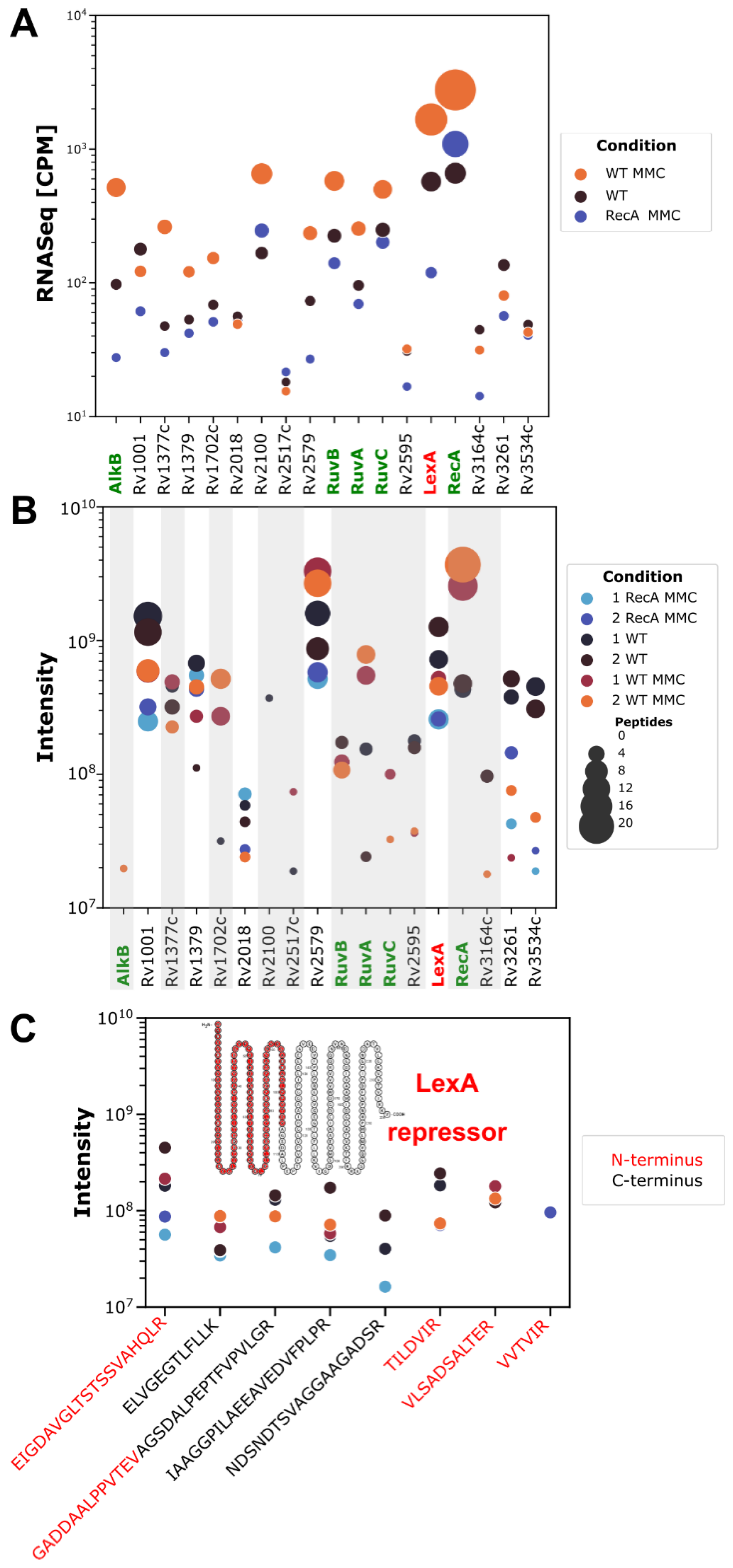
3.4. Whole-Cell Proteomics Profiling Confirms the Downregulation of the LexA Repressor in Cultures of Both Wild-Type and ΔRecA Mutant Strains Caused by RecA-Dependent Coproteolysis and Transcriptional Repression, Respectively
3.5. PafBC-Dependent Regulation of Gene Expression in Response to Mitomycin C Is Conserved among Saprophytic and Pathogenic Mycobacteria
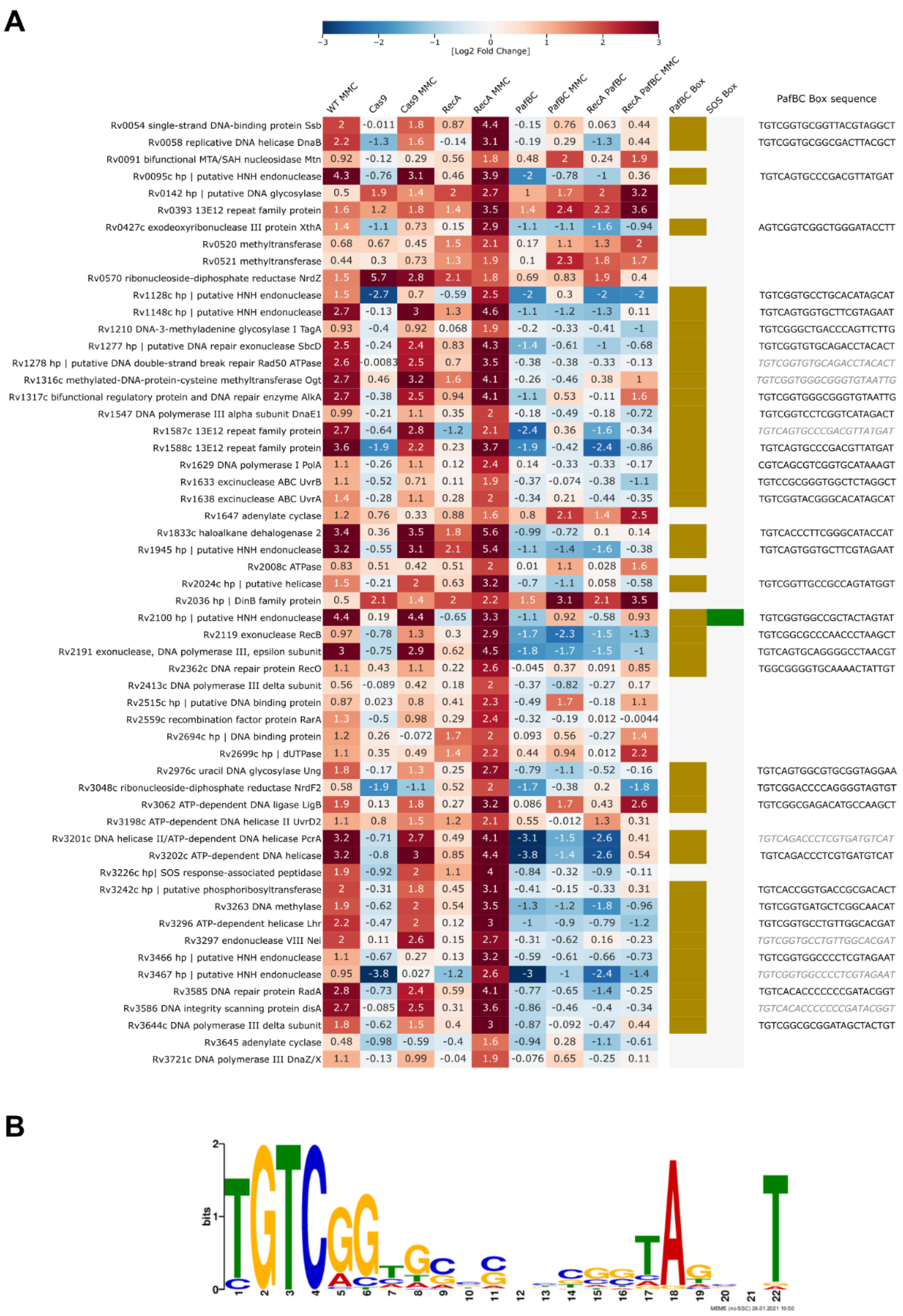
3.6. Transcriptional Profiling and Proteomics Analysis Revealed Few DNA Damage Repair Genes That Were Overexpressed in Response to DNA Damage When Both the PafBC and RecA/LexA Regulatory Networks Were Tuned Down
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020; ISBN 978-92-4-001313-1. [Google Scholar]
- Dos Vultos, T.; Mestre, O.; Tonjum, T.; Gicquel, B. DNA repair in Mycobacterium tuberculosis revisited. FEMS Microbiol. Rev. 2009, 33, 471–487. [Google Scholar] [CrossRef] [Green Version]
- Manina, G.; Griego, A.; Singh, L.K.; McKinney, J.D.; Dhar, N. Preexisting variation in DNA damage response predicts the fate of single mycobacteria under stress. EMBO J. 2019, 38, 1–19. [Google Scholar] [CrossRef]
- Płocinska, R.; Korycka-Machala, M.; Plocinski, P.; Dziadek, J. Mycobacterial DNA replication as a target for antituberculosis drug discovery. Curr. Top. Med. Chem. 2017, 17, 2129–2142. [Google Scholar] [CrossRef]
- Warner, D.F.; Ndwandwe, D.E.; Abrahams, G.L.; Kana, B.D.; Machowski, E.E.; Venclovas, Č.; Mizrahi, V. Essential roles for imuA′- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 13093–13098. [Google Scholar] [CrossRef] [Green Version]
- Castañeda-García, A.; Prieto, A.I.; Rodríguez-Beltrán, J.; Alonso, N.; Cantillon, D.; Costas, C.; Pérez-Lago, L.; Zegeye, E.D.; Herranz, M.; Plociński, P.; et al. A non-canonical mismatch repair pathway in prokaryotes. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Płociński, P.; Brissett, N.C.; Bianchi, J.; Brzostek, A.; Korycka-Machała, M.; Dziembowski, A.; Dziadek, J.; Doherty, A.J. DNA Ligase C and Prim-PolC participate in base excision repair in mycobacteria. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Shuman, S.; Glickman, M.S. RecF and RecR play critical roles in the homologous recombination and single-strand annealing pathways of mycobacteria. J. Bacteriol. 2015, 197, 3121–3132. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Unciuleac, M.-C.; Shuman, S.; Glickman, M.S. Homologous recombination mediated by the mycobacterial AdnAB helicase without end resection by the AdnAB nucleases. Nucleic Acids Res. 2017, 45, 762–774. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Patil, K.N.; Khanduja, J.S.; Kumar, P.S.; Williams, A.; Rossi, F.; Rizzi, M.; Davis, E.O.; Muniyappa, K. Mycobacterium tuberculosis UvrD1 and UvrA proteins suppress DNA strand exchange promoted by cognate and noncognate RecA proteins. Biochemistry 2010, 49, 4872–4883. [Google Scholar] [CrossRef]
- Wipperman, M.F.; Heaton, B.E.; Nautiyal, A.; Adefisayo, O.; Evans, H.; Gupta, R.; van Ditmarsch, D.; Soni, R.; Hendrickson, R.; Johnson, J.; et al. Mycobacterial mutagenesis and drug resistance are controlled by phosphorylation- and cardiolipin-mediated inhibition of the RecA coprotease. Mol. Cell 2018, 72, 152–161.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopaul, K.K.; Brooks, P.C.; Prost, J.-F.; Davis, E.O. Characterization of the two Mycobacterium tuberculosis recA promoters. J. Bacteriol. 2003, 185, 6005–6015. [Google Scholar] [CrossRef] [Green Version]
- Fudrini Olivencia, B.; Müller, A.U.; Roschitzki, B.; Burger, S.; Weber-Ban, E.; Imkamp, F. Mycobacterium smegmatis PafBC is involved in regulation of DNA damage response. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, A.U.; Imkamp, F.; Weber-Ban, E. The mycobacterial LexA/RecA-independent DNA damage response is controlled by PafBC and the pup-proteasome system. Cell Rep. 2018, 23, 3551–3564. [Google Scholar] [CrossRef]
- Brzostek, A.; Szulc, I.; Klink, M.; Brzezinska, M.; Sulowska, Z.; Dziadek, J. Either non-homologous ends joining or homologous recombination is required to repair double-strand breaks in the genome of macrophage-internalized Mycobacterium tuberculosis. PLoS ONE 2014, 9, e92799. [Google Scholar] [CrossRef]
- Rock, J.M.; Hopkins, F.F.; Chavez, A.; Diallo, M.; Chase, M.R.; Gerrick, E.R.; Pritchard, J.R.; Church, G.M.; Rubin, E.J.; Sassetti, C.M.; et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2017, 2. [Google Scholar] [CrossRef]
- Sheffield, P.; Garrard, S.; Derewenda, Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999, 15, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Korycka-Machała, M.; Pawełczyk, J.; Borówka, P.; Dziadek, B.; Brzostek, A.; Kawka, M.; Bekier, A.; Rykowski, S.; Olejniczak, A.B.; Strapagiel, D.; et al. PPE51 Is involved in the uptake of disaccharides by Mycobacterium tuberculosis. Cells 2020, 9, 603. [Google Scholar] [CrossRef] [Green Version]
- Korycka-Machala, M.; Rychta, E.; Brzostek, A.; Sayer, H.R.; Rumijowska-Galewicz, A.; Bowater, R.P.; Dziadek, J. Evaluation of NAD(+)-dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob. Agents Chemother. 2007, 51, 2888–2897. [Google Scholar] [CrossRef] [Green Version]
- Pawelczyk, J.; Brzostek, A.; Kremer, L.; Dziadek, B.; Rumijowska-Galewicz, A.; Fiolka, M.; Dziadek, J. AccD6, a key carboxyltransferase essential for mycolic acid synthesis in Mycobacterium tuberculosis, is dispensable in a nonpathogenic strain. J. Bacteriol. 2011, 193, 6960–6972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Płociński, P.; Macios, M.; Houghton, J.; Niemiec, E.; Płocińska, R.; Brzostek, A.; Słomka, M.; Dziadek, J.; Young, D.; Dziembowski, A. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 2019, 47, 5892–5905. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Powell, D. Degust: Powerfull and User Friendly Front-End Data Analsysis, Visualisation and Exploratory Tool for RNA-Sequencing. Available online: https://github.com/drpowell/degust (accessed on 22 March 2021).
- Góralczyk-Bińkowska, A.; Jasińska, A.; Długoński, A.; Płociński, P.; Długoński, J. Laccase activity of the ascomycete fungus Nectriella pironii and innovative strategies for its production on leaf litter of an urban park. PLoS ONE 2020, 15, e0231453. [Google Scholar]
- Minias, A.; Minias, P.; Czubat, B.; Dziadek, J. Purifying selective pressure suggests the functionality of a vitamin B12 biosynthesis pathway in a global population of Mycobacterium tuberculosis. Genome Biol. Evol. 2018, 10, 2326–2337. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Korycka-Machala, M.; Brzostek, A.; Rozalska, S.; Rumijowska-Galewicz, A.; Dziedzic, R.; Bowater, R.; Dziadek, J. Distinct DNA repair pathways involving RecA and nonhomologous end joining in Mycobacterium smegmatis. FEMS Microbiol. Lett. 2006, 258, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Smollett, K.L.; Smith, K.M.; Kahramanoglou, C.; Arnvig, K.B.; Buxton, R.S.; Davis, E.O. Global analysis of the regulon of the transcriptional repressor LexA, a key component of SOS response in Mycobacterium tuberculosis. J. Biol. Chem. 2012, 287, 22004–22014. [Google Scholar] [CrossRef] [Green Version]
- Rand, L.; Hinds, J.; Springer, B.; Sander, P.; Buxton, R.S.; Davis, E.O. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 2003, 50, 1031–1042. [Google Scholar] [CrossRef]
- Dawson, L.F.; Dillury, J.; Davis, E.O. RecA-Independent DNA damage induction of Mycobacterium tuberculosis ruvC despite an appropriately located SOS box. J. Bacteriol. 2010, 192, 599–603. [Google Scholar] [CrossRef] [Green Version]
- Minch, K.J.; Rustad, T.R.; Peterson, E.J.R.; Winkler, J.; Reiss, D.J.; Ma, S.; Hickey, M.; Brabant, W.; Morrison, B.; Turkarslan, S.; et al. The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yellaboina, S.; Ranjan, S.; Vindal, V.; Ranjan, A. Comparative analysis of iron regulated genes in mycobacteria. FEBS Lett. 2006, 580, 2567–2576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, G.M.; Voskuil, M.I.; Gold, B.; Schoolnik, G.K.; Smith, I. ideR, an essential gene in Mycobacterium tuberculosis: Role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 2002, 70, 3371–3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Jiang, X.; Xu, M.; Fang, Y.; Wang, Y.; Sun, G.; Guo, J. Identification of stress-responsive transcription factors with protein-bound Escherichia coli genomic DNA libraries. AMB Express 2020, 10. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Jiang, J.; Ng, M.; Cui, Z.; Mai, J.; Ahn, S.K.; Liu, J.; Zhang, J.; Liu, J.; et al. Transcription factors Rv0081 and Rv3334 connect the early and the enduring hypoxic response of Mycobacterium tuberculosis. Virulence 2018, 9, 1468–1482. [Google Scholar] [CrossRef] [Green Version]
- Davis, E.O.; Dullaghan, E.M.; Rand, L. Definition of the mycobacterial SOS box and use to identify LexA-regulated genes in Mycobacterium tuberculosis. J. Bacteriol. 2002, 184, 3287–3295. [Google Scholar] [CrossRef] [Green Version]
- Mo, C.Y.; Birdwell, L.D.; Kohli, R.M. Specificity determinants for autoproteolysis of LexA, a key regulator of bacterial SOS mutagenesis. Biochemistry 2014, 53, 3158–3168. [Google Scholar] [CrossRef]
- Liu, J.; Ehmsen, K.T.; Heyer, W.-D.; Morrical, S.W. Presynaptic filament dynamics in homologous recombination and DNA repair. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 240–270. [Google Scholar] [CrossRef] [Green Version]
- Gataulin, D.V.; Carey, J.N.; Li, J.; Shah, P.; Grubb, J.T.; Bishop, D.K. The ATPase activity of E. coli RecA prevents accumulation of toxic complexes formed by erroneous binding to undamaged double stranded DNA. Nucleic Acids Res. 2018, 46, 9510–9523. [Google Scholar] [CrossRef] [Green Version]
- Zahradka, K.; Buljubašić, M.; Petranović, M.; Zahradka, D. Roles of ExoI and SbcCD nucleases in “reckless” DNA degradation in recA mutants of Escherichia coli. J. Bacteriol. 2009, 191, 1677–1687. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.L.; Cox, M.M.; Inman, R.B. Recombinational DNA repair: The RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 1997, 91, 347–356. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Ryzhikov, M.; Koroleva, O.; Unciuleac, M.; Shuman, S.; Korolev, S.; Glickman, M.S. A dual role for mycobacterial RecO in RecA-dependent homologous recombination and RecA-independent single-strand annealing. Nucleic Acids Res. 2013, 41, 2284–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dullaghan, E.M.; Brooks, P.C.; Davis, E.O. The role of multiple SOS boxes upstream of the Mycobacterium tuberculosis lexA gene—identification of a novel DNA-damage-inducible gene. Microbiology 2002, 148, 3609–3615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Yang, R.; Lyu, M.; Wang, S.; Liu, X.; Wen, Y.; Song, Y.; Li, J.; Chen, Z. IdeR, a DtxR family iron response regulator, controls iron homeostasis, morphological differentiation, secondary metabolism, and the oxidative stress response in Streptomyces avermitilis. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, R.; Rodriguez, G.M. IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol. Microbiol. 2014, 91, 98–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.-N.; Lee, N.-O.; Han, S.J.; Ko, I.-J.; Oh, J.-I. Regulation of the ahpC gene encoding alkyl hydroperoxide reductase in Mycobacterium smegmatis. PLoS ONE 2014, 9, e111680. [Google Scholar] [CrossRef]
- Rustad, T.R.; Minch, K.J.; Ma, S.; Winkler, J.K.; Hobbs, S.; Hickey, M.; Brabant, W.; Turkarslan, S.; Price, N.D.; Baliga, N.S.; et al. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 2014, 15. [Google Scholar] [CrossRef]
- Iacobino, A.; Piccaro, G.; Pardini, M.; Fattorini, L.; Giannoni, F. Moxifloxacin activates the sos response in Mycobacterium tuberculosis in a dose-and time-dependent manner. Microorganisms 2021, 9, 255. [Google Scholar] [CrossRef]
- Jagielski, T.; Bakuła, Z.; Brzostek, A.; Minias, A.; Stachowiak, R.; Kalita, J.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Żaczek, A.; Vasiliauskiene, E.; et al. Characterization of mutations conferring resistance to rifampin in Mycobacterium tuberculosis clinical strains. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Gough, J. Convergent evolution of domain architectures (is rare). Bioinformatics 2005, 21, 1464–1471. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brzostek, A.; Płociński, P.; Minias, A.; Ciszewska, A.; Gąsior, F.; Pawełczyk, J.; Dziadek, B.; Słomka, M.; Dziadek, J. Dissecting the RecA-(In)dependent Response to Mitomycin C in Mycobacterium tuberculosis Using Transcriptional Profiling and Proteomics Analyses. Cells 2021, 10, 1168. https://doi.org/10.3390/cells10051168
Brzostek A, Płociński P, Minias A, Ciszewska A, Gąsior F, Pawełczyk J, Dziadek B, Słomka M, Dziadek J. Dissecting the RecA-(In)dependent Response to Mitomycin C in Mycobacterium tuberculosis Using Transcriptional Profiling and Proteomics Analyses. Cells. 2021; 10(5):1168. https://doi.org/10.3390/cells10051168
Chicago/Turabian StyleBrzostek, Anna, Przemysław Płociński, Alina Minias, Aneta Ciszewska, Filip Gąsior, Jakub Pawełczyk, Bożena Dziadek, Marcin Słomka, and Jarosław Dziadek. 2021. "Dissecting the RecA-(In)dependent Response to Mitomycin C in Mycobacterium tuberculosis Using Transcriptional Profiling and Proteomics Analyses" Cells 10, no. 5: 1168. https://doi.org/10.3390/cells10051168





