Genetic Perturbation of Pyruvate Dehydrogenase Kinase 1 Modulates Growth, Angiogenesis and Metabolic Pathways in Ovarian Cancer Xenografts
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
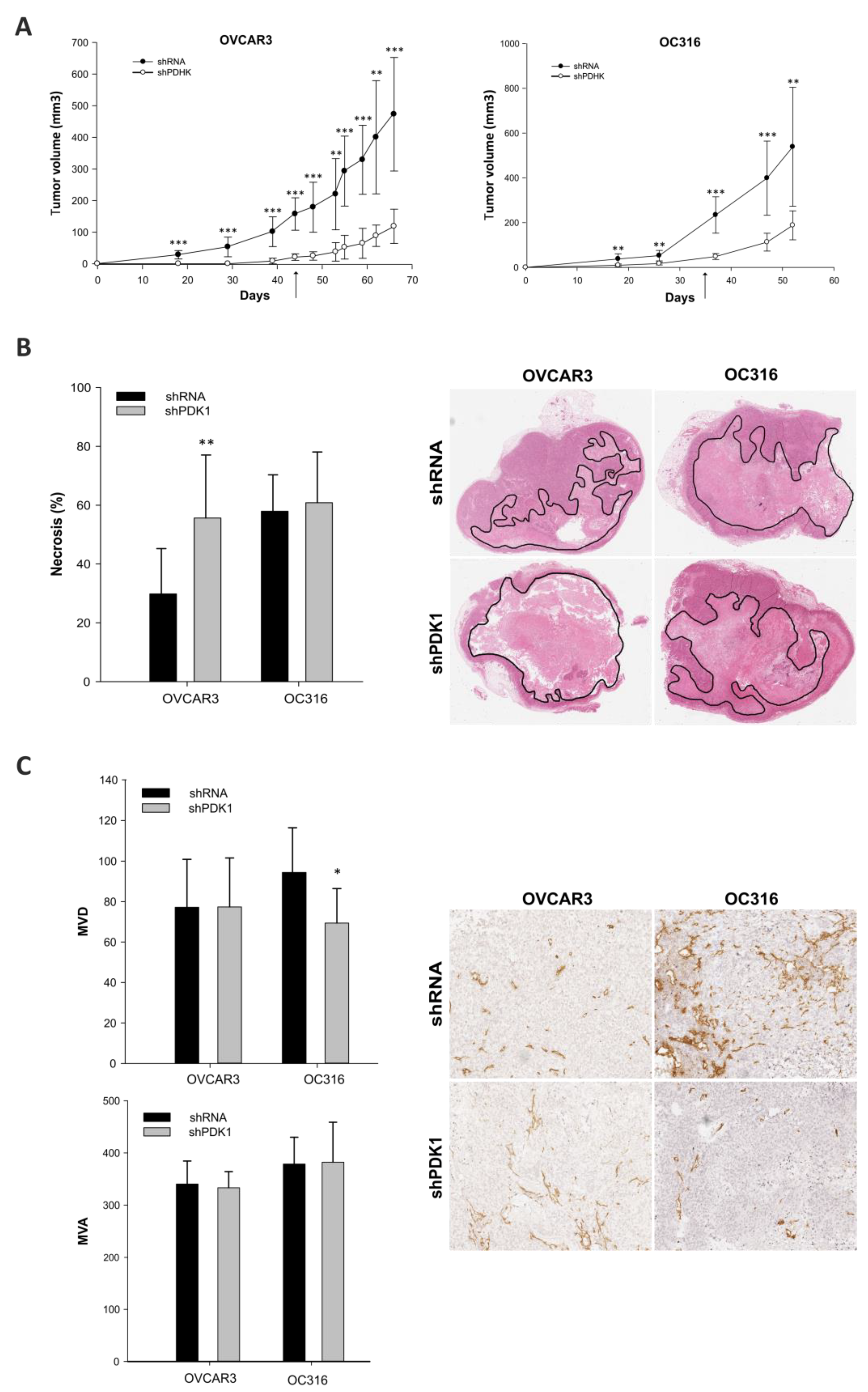
2.2. In Vivo Experiments
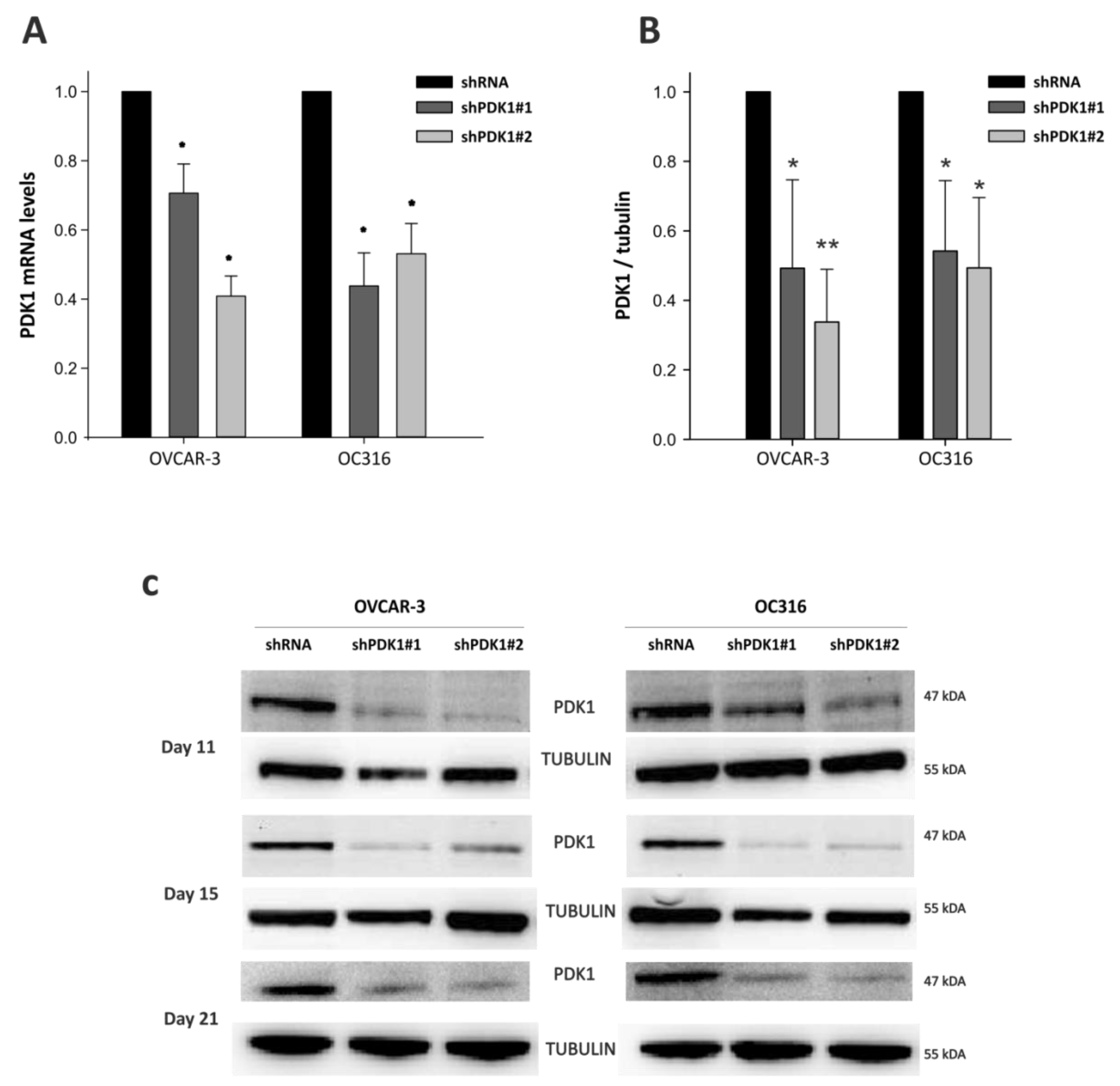
2.3. Lentiviral Vector-Mediated PDK1 Gene Silencing
2.4. Reverse Transcription-PCR (RT-PCR) and Quantitative PCR (qPCR)
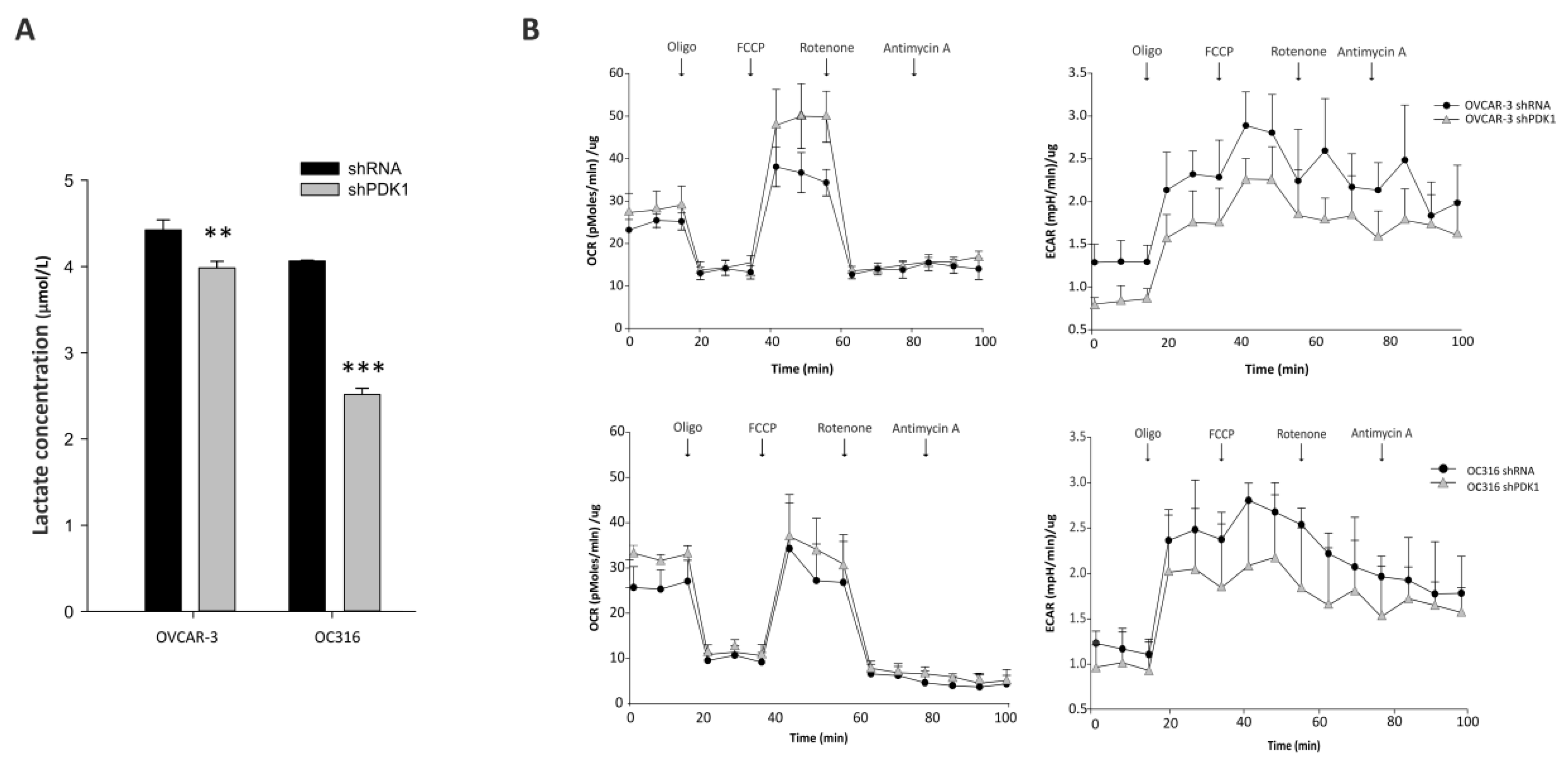
2.5. Lactate Measurements
2.6. Seahorse Analysis
2.7. Proliferation Assay
2.8. PKH26 Staining
2.9. Immunoblotting Assay
2.10. Histology and Immunohistochemistry
2.11. Image Acquisition and Analysis
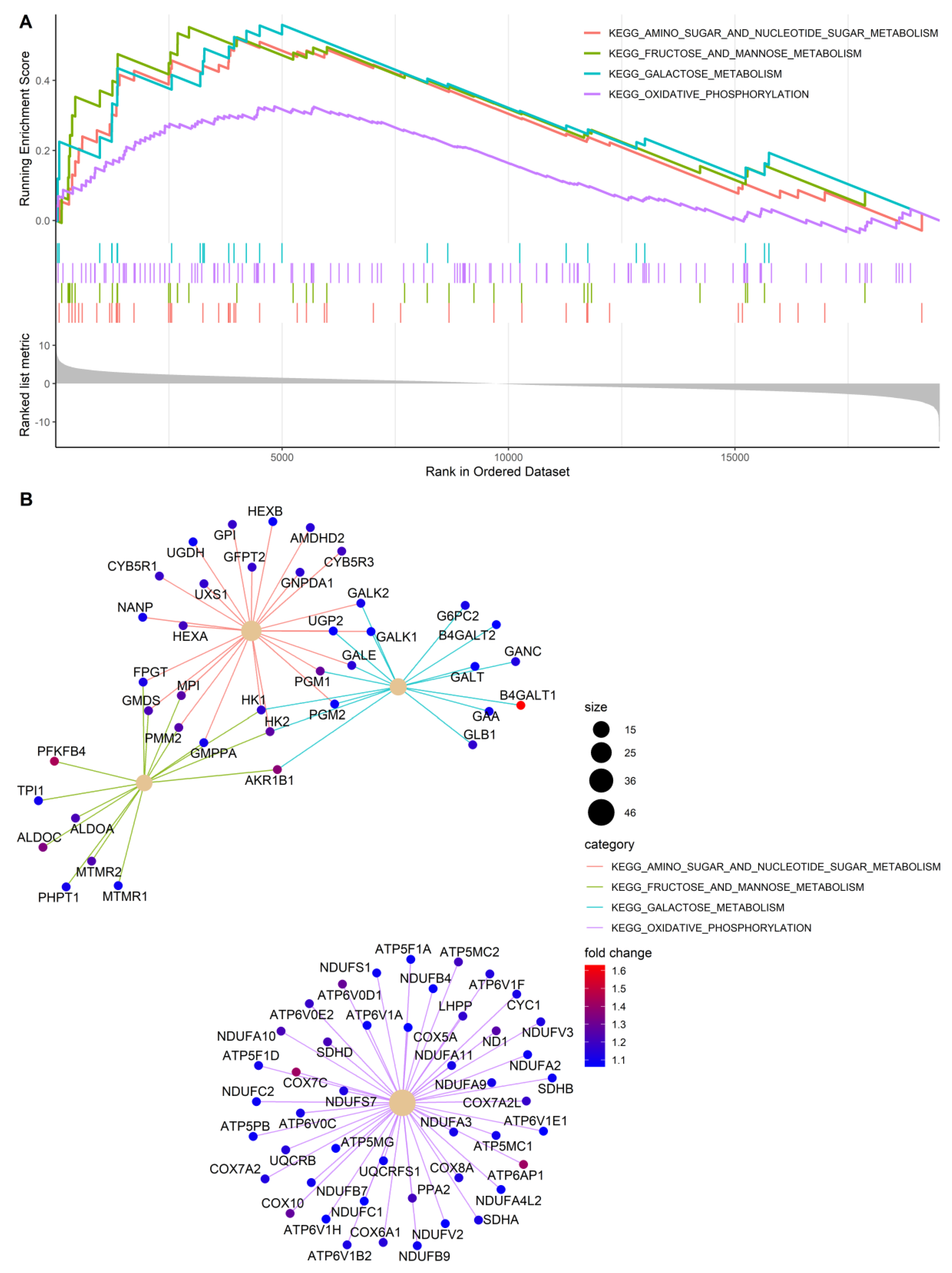
2.12. Microarray Expression Analysis
2.13. Statistical Analysis
3. Results
3.1. PDK Silencing Modulates Glycolysis and In Vitro Growth of Ovarian Cancer Cells
3.2. Effects of PDK Silencing on Tumor Growth and Angiogenesis
3.3. Gene Expression Profiles of PDK-Silenced Tumors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Allal, A.S.; Dulguerov, P.; Allaoua, M.; Haenggeli, C.A.; El-Ghazi el, A.; Lehmann, W.; Slosman, D.O. Standardized uptake value of 2-[(18)F] fluoro-2-deoxy-D-glucose in predicting outcome in head and neck carcinomas treated by radiotherapy with or without chemotherapy. J. Clin. Oncol. 2002, 20, 1398–1404. [Google Scholar] [CrossRef]
- Choi, E.S.; Ha, S.G.; Kim, H.S.; Ha, J.H.; Paeng, J.C.; Han, I. Total lesion glycolysis by 18F-FDG PET/CT is a reliable predictor of prognosis in soft-tissue sarcoma. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 1836–1842. [Google Scholar] [CrossRef]
- Chung, H.W.; Lee, K.Y.; Kim, H.J.; Kim, W.S.; So, Y. FDG PET/CT metabolic tumor volume and total lesion glycolysis predict prognosis in patients with advanced lung adenocarcinoma. J. Cancer Res. Clin. Oncol. 2014, 140, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Cho, A.; Lee, J.H.; Yun, M.; Lee, J.D.; Kim, Y.T.; Kang, W.J. The role of metabolic tumor volume and total lesion glycolysis on (1)(8)F-FDG PET/CT in the prognosis of epithelial ovarian cancer. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1898–1906. [Google Scholar] [CrossRef] [PubMed]
- Walenta, S.; Mueller-Klieser, W.F. Lactate: Mirror and motor of tumor malignancy. Semin. Radiat. Oncol. 2004, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Walenta, S.; Schroeder, T.; Mueller-Klieser, W. Lactate in solid malignant tumors: Potential basis of a metabolic classification in clinical oncology. Curr. Med. Chem. 2004, 11, 2195–2204. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Sonveaux, P.; Copetti, T.; De Saedeleer, C.J.; Vegran, F.; Verrax, J.; Kennedy, K.M.; Moon, E.J.; Dhup, S.; Danhier, P.; Frerart, F.; et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS ONE 2012, 7, e33418. [Google Scholar]
- Martinez-Outschoorn, U.E.; Pavlides, S.; Howell, A.; Pestell, R.G.; Tanowitz, H.B.; Sotgia, F.; Lisanti, M.P. Stromal-epithelial metabolic coupling in cancer: Integrating autophagy and metabolism in the tumor microenvironment. Int. J. Biochem. Cell Biol. 2011, 43, 1045–1051. [Google Scholar] [CrossRef]
- Vegran, F.; Boidot, R.; Michiels, C.; Sonveaux, P.; Feron, O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-kappaB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011, 71, 2550–2560. [Google Scholar] [CrossRef]
- Payen, V.L.; Mina, E.; Van Hee, V.F.; Porporato, P.E.; Sonveaux, P. Monocarboxylate transporters in cancer. Mol. Metab. 2019, 33, 48–66. [Google Scholar] [CrossRef]
- Piga, I.; Verza, M.; Montenegro, F.; Nardo, G.; Zulato, E.; Zanin, T.; Del Bianco, P.; Esposito, G.; Indraccolo, S. In situ Metabolic Profiling of Ovarian Cancer Tumor Xenografts: A Digital Pathology Approach. Front. Oncol. 2020, 10, 1277. [Google Scholar] [CrossRef]
- Bonnet, S.; Archer, S.L.; Allalunis-Turner, J.; Haromy, A.; Beaulieu, C.; Thompson, R.; Lee, C.T.; Lopaschuk, G.D.; Puttagunta, L.; Harry, G.; et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 2007, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Tataranni, T.; Piccoli, C. Dichloroacetate (DCA) and Cancer: An Overview towards Clinical Applications. Oxid. Med. Cell Longev. 2019, 2019, 8201079. [Google Scholar] [CrossRef] [PubMed]
- Sutendra, G.; Dromparis, P.; Kinnaird, A.; Stenson, T.H.; Haromy, A.; Parker, J.M.; McMurtry, M.S.; Michelakis, E.D. Mitochondrial activation by inhibition of PDKII suppresses HIF1a signaling and angiogenesis in cancer. Oncogene 2013, 32, 1638–1650. [Google Scholar] [CrossRef]
- Mayers, J.R.; Vander Heiden, M.G. Famine versus feast: Understanding the metabolism of tumors in vivo. Trends Biochem. Sci. 2015, 40, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Fabian, C.; Koetz, L.; Favaro, E.; Indraccolo, S.; Mueller-Klieser, W.; Sattler, U.G. Protein profiles in human ovarian cancer cell lines correspond to their metabolic activity and to metabolic profiles of respective tumor xenografts. FEBS J. 2012, 279, 882–891. [Google Scholar] [CrossRef]
- Nardo, G.; Favaro, E.; Curtarello, M.; Moserle, L.; Zulato, E.; Persano, L.; Rossi, E.; Esposito, G.; Crescenzi, M.; Casanovas, O.; et al. Glycolytic phenotype and AMP kinase modify the pathologic response of tumor xenografts to VEGF neutralization. Cancer Res. 2011, 71, 4214–4225. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Pasto, A.; Marchesi, M.; Diamantini, A.; Frasson, C.; Curtarello, M.; Lago, C.; Pilotto, G.; Parenti, A.R.; Esposito, G.; Agostini, M.; et al. PKH26 staining defines distinct subsets of normal human colon epithelial cells at different maturation stages. PLoS ONE 2012, 7, e43379. [Google Scholar] [CrossRef]
- Zhang, M.; Cong, Q.; Zhang, X.Y.; Zhang, M.X.; Lu, Y.Y.; Xu, C.J. Pyruvate dehydrogenase kinase 1 contributes to cisplatin resistance of ovarian cancer through EGFR activation. J. Cell Physiol. 2019, 234, 6361–6370. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Ni, Y.; Chen, F. Tumor resistance to vascular disrupting agents: Mechanisms, imaging, and solutions. Oncotarget 2016, 7, 15444–15459. [Google Scholar] [CrossRef]
- Beaufort, C.M.; Helmijr, J.C.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van Ijcken, W.F.; Heine, A.A.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Tudrej, P.; Kujawa, K.A.; Cortez, A.J.; Lisowska, K.M. Characteristics of in Vivo Model Systems for Ovarian Cancer Studies. Diagnostics 2019, 9, 120. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef]
- Dupuy, F.; Tabaries, S.; Andrzejewski, S.; Dong, Z.; Blagih, J.; Annis, M.G.; Omeroglu, A.; Gao, D.; Leung, S.; Amir, E.; et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015, 22, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Yin, H. PDK1 promotes tumor cell proliferation and migration by enhancing the Warburg effect in non-small cell lung cancer. Oncol. Rep. 2017, 37, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, J.H.; Fan, W.J.; Meng, Y.T.; Li, M.M.; Li, T.T.; Cui, B.; Wang, H.F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2018, 37, 1062–1074. [Google Scholar] [CrossRef]
- Semba, H.; Takeda, N.; Isagawa, T.; Sugiura, Y.; Honda, K.; Wake, M.; Miyazawa, H.; Yamaguchi, Y.; Miura, M.; Jenkins, D.M.; et al. HIF-1alpha-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat. Commun. 2016, 7, 11635. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venturoli, C.; Piga, I.; Curtarello, M.; Verza, M.; Esposito, G.; Venuto, S.; Navaglia, F.; Grassi, A.; Indraccolo, S. Genetic Perturbation of Pyruvate Dehydrogenase Kinase 1 Modulates Growth, Angiogenesis and Metabolic Pathways in Ovarian Cancer Xenografts. Cells 2021, 10, 325. https://doi.org/10.3390/cells10020325
Venturoli C, Piga I, Curtarello M, Verza M, Esposito G, Venuto S, Navaglia F, Grassi A, Indraccolo S. Genetic Perturbation of Pyruvate Dehydrogenase Kinase 1 Modulates Growth, Angiogenesis and Metabolic Pathways in Ovarian Cancer Xenografts. Cells. 2021; 10(2):325. https://doi.org/10.3390/cells10020325
Chicago/Turabian StyleVenturoli, Carolina, Ilaria Piga, Matteo Curtarello, Martina Verza, Giovanni Esposito, Santina Venuto, Filippo Navaglia, Angela Grassi, and Stefano Indraccolo. 2021. "Genetic Perturbation of Pyruvate Dehydrogenase Kinase 1 Modulates Growth, Angiogenesis and Metabolic Pathways in Ovarian Cancer Xenografts" Cells 10, no. 2: 325. https://doi.org/10.3390/cells10020325
APA StyleVenturoli, C., Piga, I., Curtarello, M., Verza, M., Esposito, G., Venuto, S., Navaglia, F., Grassi, A., & Indraccolo, S. (2021). Genetic Perturbation of Pyruvate Dehydrogenase Kinase 1 Modulates Growth, Angiogenesis and Metabolic Pathways in Ovarian Cancer Xenografts. Cells, 10(2), 325. https://doi.org/10.3390/cells10020325






