The Role of Non-Canonical Hsp70s (Hsp110/Grp170) in Cancer
Abstract
:1. Introduction
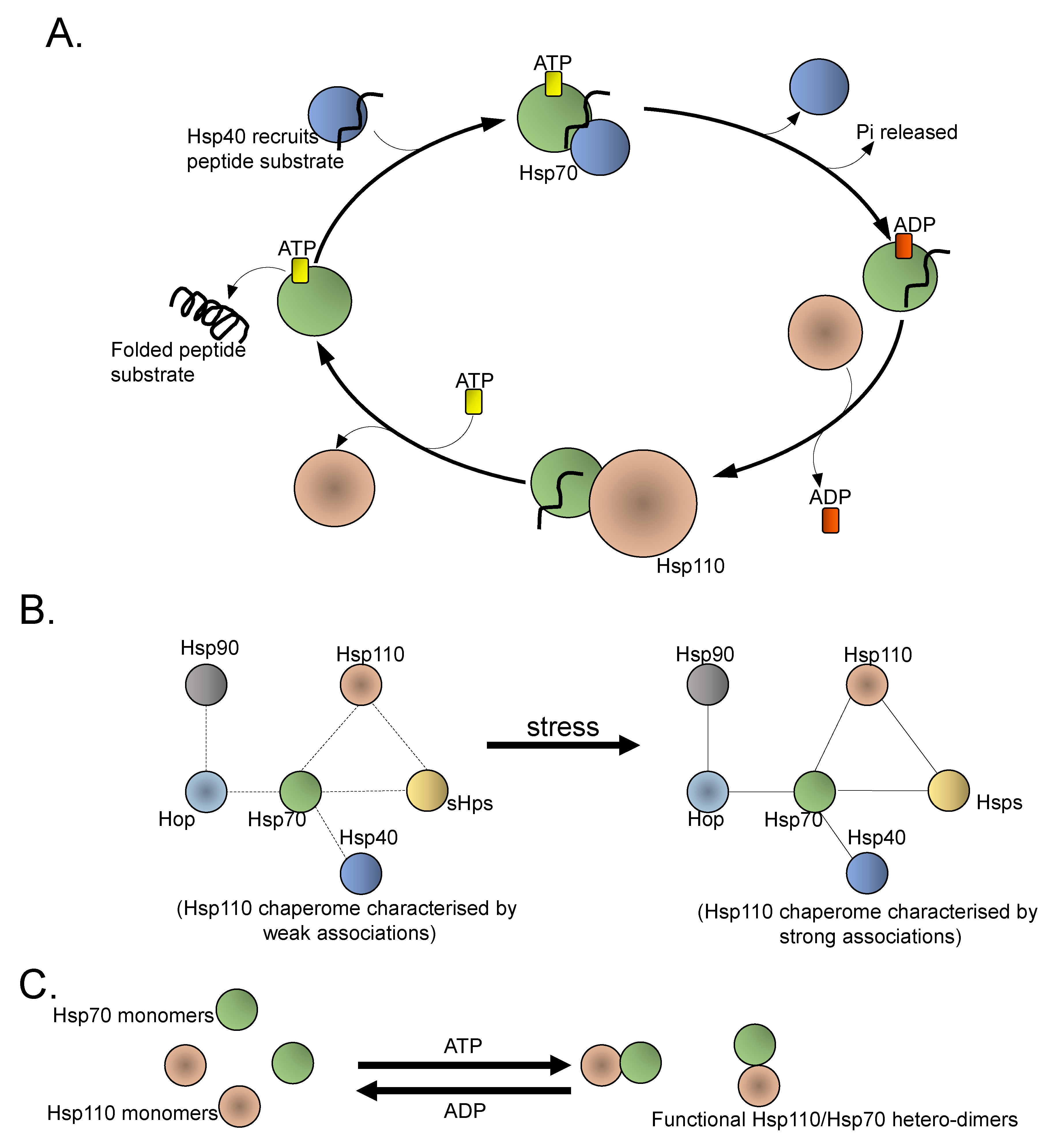
2. Hsp110/Grp170
3. Hsp110 Roles in Cancer Pathogenesis
4. The Role of the ER Resident, Grp170 Chaperone in Cancer Pathophysiology
5. Identification of Potential Unique Proteomic Signatures of Hsp110/Grp170
6. Heroes or Villains: The Role of Heat Shock Proteins in Preventing Cancer Progression
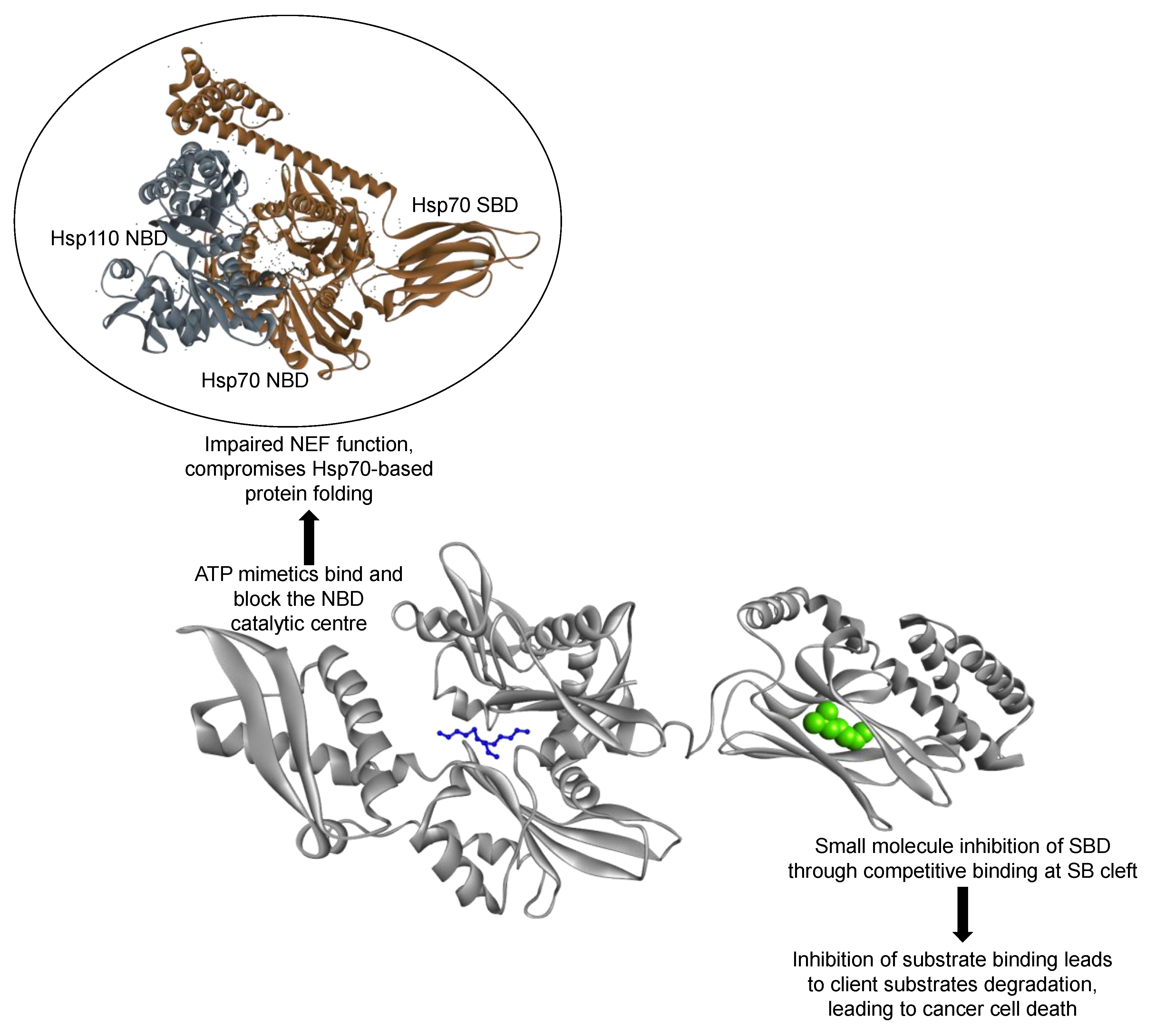
7. Could Hsp110/Grp170 Be Targeted in Cancer Therapy?
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report: Cancer Research for Cancer Prevention; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Arruebo, M.; Vilaboa, N.; Saez, B.; Lambea, J.; Tres, A.; Valladares, M.; González-Fernández, Á. Assessment of the evolution of cancer treatment therapies. Cancers 2011, 3, 3279–3330. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Rodger, E.J.; Eccles, M. Epigenetic drivers of tumorigenesis and cancer metastasis. Semin. Cancer Biol. 2018, 51, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Klimczak, M.; Biecek, P.; Zylicz, A.; Zylicz, M. Heat shock proteins create a signature to predict the clinical outcome in breast cancer. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, C.; Manero, F.; Gonin, S.; Kretz-Remy, C.; Virot, S.; Arrigo, A.P. Hsp27 as a negative regulator of cytochrome C release. Mol. Cell Biol. 2002, 22, 816–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, V.P.; Jain, R.K. Strategies for advancing cancer nanomedicine. Nat. Mater. 2013, 12, 958–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, O.H.; Batra, S.; Kolattukudy, S.J.; Gonzalez-Mejia, M.E.; Smith, J.B.; Doseff, A.I. Binding of caspase-3 prodomain to heat shock protein 27 regulates monocyte apoptosis by inhibiting caspase-3 proteolytic activation. J. Biol. Chem. 2007, 282, 25088–25099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acunzo, J.; Katsogiannou, M.; Rocchi, P. Small heat shock proteins HSP27 (HspB1), αB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int. J. Biochem. Cell Biol. 2012, 44, 1622–1631. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Zhou, Y.; Wang, Y.; Wang, S.; Zhang, W. Hsp70 promotes chemoresistance by blocking Bax mitochondrial translocation in ovarian cancer cells. Cancer Lett. 2012, 321, 137–143. [Google Scholar] [CrossRef]
- Stankiewicz, A.R.; Lachapelle, G.; Foo, C.P.Z.; Radicioni, S.M.; Mosser, D.D. Hsp70 inhibits heat-induced apoptosis ppstream of mitochondria by preventing Bax translocation. J. Biol. Chem. 2005, 280, 38729–38739. [Google Scholar] [CrossRef] [Green Version]
- Ischia, J.; So, A.I. The role of heat shock proteins in bladder cancer. Nat. Rev. Urol. 2013, 10, 386–395. [Google Scholar] [CrossRef]
- Ferreira, L.M.R.; Cunha-Oliveira, T.; Sobral, M.C.; Abreu, P.L.; Alpoim, M.C.; Urbano, A.M. Impact of carcinogenic chromium on the cellular response to proteotoxic stress. Int. J. Mol. Sci. 2019, 20, 4901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Khaleque, M.A.; Sawyer, D.B.; Ciocca, D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Nahleh, Z.; Tfayli, A.; Najm, A.; El Sayed, A.; Nahle, Z. Heat shock proteins in cancer: Targeting the ‘chaperones’. Future Med. Chem. 2012, 4, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Dimas, D.T.; Perlepe, C.D.; Sergentanis, T.N.; Misitzis, I.; Kontzoglou, K.; Patsouris, E.; Kouraklis, G.; Psaltopoulou, T.; Nonni, A. The prognostic significance of Hsp70/Hsp90 expression in breast cancer: A systematic review and meta-analysis. Anticancer Res. 2018, 38, 1551–1562. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Stevenson, M.A.; Murshid, A. Heat shock proteins, autoimmunity, and aancer treatment. Autoimmune Dis. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yun, C.W.; Kim, H.J.; Lim, J.H.; Lee, S.H. Heat shock proteins: Agents of cancer development and therapeutic targets in anticancer therapy. Cells 2019, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Fan, G.; Tu, Y.; Wu, N.; Xiao, H. The expression profiles and prognostic values of HSPs family members in head and neck cancer. Cancer Cell Int. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Gopal, A.; Gopal, U. Cell Surface Heart shock proteins and their role in cancer. Clin. Pathol. 2017, 1, 000104. [Google Scholar] [CrossRef]
- Zininga, T.; Ramatsui, L.; Shonhai, A. Heat shock proteins as immunomodulants. Molecules 2018, 23, 2846. [Google Scholar] [CrossRef] [Green Version]
- Botzler, C.; Li, G.; Issels, R.D.; Multhoff, G. Definition of extracellular localized epitopes of Hsp70 involved in an NK immune response. Cell Stress Chaperones 1998, 3, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Multhoff, G.; Mizzen, L.; Winchester, C.C.; Milner, C.M.; Wenk, S.; Eissner, G.; Kampinga, H.H.; Laumbacher, B.; Johnson, J. Heat shock protein 70 (Hsp70) stimulates proliferation and cytolytic activity of natural killer cells. Exp. Hematol. 1999, 27, 1627–1636. [Google Scholar] [CrossRef]
- Wu, X.; Wanders, A.; Wardega, P.; Tinge, B.; Gedda, L.; Bergstrom, S.M.; Sooman, L.; Gullbo, J.; Bergqvist, M.; Hesselius, P.; et al. Hsp90 is expressed and represents a therapeutic target in human oesophageal cancer using the inhibitor 17-allylamino-17-demethoxygeldanamycin. Br. J. Cancer 2009, 100, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Garg, G.; Khandelwal, A.; Blagg, B.S. Anticancer inhibitors of Hsp90 function: Beyond the usual suspects. Adv. Cancer Res. 2016, 129, 51–88. [Google Scholar] [PubMed] [Green Version]
- Chavany, C.; Mimnaugh, E.; Miller, P.; Bitton, R.; Nguyen, P.; Trepel, J.; Whitesell, L.; Schnur, R.; Moyer, J.; Neckers, L. p185erbB2 binds to GRP94 in vivo. Dissociation of the p185erbB2/GRP94 heterocomplex by benzoquinone ansamycins pre-cedes depletion of p185erbB2. Biol. Chem. 1996, 9, 4974–4977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Hagn, F.; Lagleder, S.; Retzlaff, M.; Rohrberg, J.; Demmer, O.; Richter, K.W.; Buchner, J.; Kessler, H. Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nat. Struct. Mol. Biol. 2011, 18, 1086–1093. [Google Scholar] [CrossRef]
- Kudryavtsev, V.A.; Khokhlova, A.V.; Mosina, V.A.; Selivanova, E.I.; Kabakov, A.E. Induction of Hsp70 in tumor cells treated with inhibitors of the Hsp90 activity: A predictive marker and promising target for radiosensitization. PLoS ONE 2017, 12, e0173640. [Google Scholar] [CrossRef]
- Beere, H.M.; Wolf, B.B.; Cain, K.; Mosser, D.D.; Mahboubi, A.; Kuwana, T.; Tailor, P.; Morimoto, R.I.; Cohen, G.M.; Green, D.R. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2000, 2, 469–475. [Google Scholar] [CrossRef]
- Ikwegbue, P.C.; Masamba, P.; Oyinloye, B.E.; Kappo, A.P. Roles of heat shock proteins in apoptosis, oxidative stress, human inflammatory diseases, and cancer. Pharmaceuticals 2018, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- Venugopal, A.; Stoffel, E.M. Colorectal cancer in young adults. Curr. Treat. Opt. Gastroenterol. 2019, 17, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.D.; Morimoto, R.I. Molecular chaperones and the stress of oncogenesis. Oncogene 2004, 23, 2907–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, M.Y.; Gabai, V.L. Hsp70 in cancer: Back to the future. Oncogene 2015, 34, 4153–4161. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Chen, J.; Loo, A.; Jaeger, S.; Bagdasarian, L.; Yu, J.; Chung, F.; Korn, J.; Ruddy, D.; Guo, R.; et al. Targeting HSF1 sensitizes cancer cells to HSP90 inhibition. Oncotarget 2013, 4, 816–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Chen, M.; Zhou, J.; Zhang, X. HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy. Int. J. Oncol. 2014, 45, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Burns, T.F. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaglom, J.A.; Wang, Y.; Li, A.; Li, Z.; Monti, S.; Alexandrov, I.; Lu, X.; Sherman, M.Y. Cancer cell responses to Hsp70 inhibitor JG-98: Comparison with Hsp90 inhibitors and finding synergistic drug combinations. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yin, F.; Feng, F.; Wang, L.; Wang, X.; Li, Z.; Cao, Y. SREBP-1 inhibitor Betulin enhances the antitumor effect of Sorafenib on hepatocellular carcinoma via restricting cellular glycolytic activity. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2008, 14, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Easton, D.P.; Kaneko, Y.; Subjeck, J.R. The Hsp110 and Grp170 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones 2000, 5, 276–290. [Google Scholar] [CrossRef] [Green Version]
- Chakafana, G.; Zininga, T.; Shonhai, A. Comparative structure-function features of Hsp70s of Plasmodium falciparum and human origins. Biophys. Rev. 2019, 11, 591–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, H.J.; Easton, D.; Murawski, M.; Kaneko, Y.; Subjeck, J.R. The chaperoning activity of hsp110 identification of functional domains by use of targeted deletions. J. Biol. Chem. 1999, 274, 15712–15718. [Google Scholar] [CrossRef] [Green Version]
- Zininga, T.; Achilonu, I.; Hoppe, H.C.; Prinsloo, E.; Dirr, H.W.; Shonhai, A. Plasmodium falciparum Hsp70-z, an Hsp110 homologue, exhibits independent chaperone activity and interacts with Hsp70-1 in a nucleotide-dependent fashion. Cell Stress Chaperones 2016, 21, 499–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakafana, G.; Zininga, T.; Shonhai, A. The link that binds: The linker of Hsp70 as a helm of the protein’s function. Biomolecules 2019, 9, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zappasodi, R.; Ruggiero, G.; Guarnotta, C.; Tortoreto, M.; Tringali, C.; Cavanè, A.; Cabras, A.D.; Castagnoli, L.; Venerando, B.; Zaffaroni, N.; et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood 2015, 125, 1768–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, H.; Furukawa, T.; Yano, T.; Sato, N.; Takizawa, J.; Kurasaki, T.; Abe, T.; Narita, M.; Masuko, M.; Koyama, S.; et al. Identification of an overexpressed gene, HSPA4L, the product of which can provoke prevalent humoral immune responses in leukemia patients. Exp. Hematol. 2007, 35, 1091–1099. [Google Scholar] [CrossRef]
- Held, T.; Barakat, A.Z.; Mohamed, B.A.; Paprotta, I.; Meinhardt, A.; Engel, W.; Adham, I.M. Heat-shock protein HSPA4 is required for progression of spermatogenesis. Reproduction 2011, 142, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Behnke, J.; Mann, M.J.; Scruggs, F.-L.; Feige, M.J.; Hendershot, L.M. Members of the Hsp70 Family Recognize Distinct Types of Sequences to Execute ER Quality Control. Mol. Cell 2016, 63, 739–752. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Sarbeng, E.B.; Vorvis, C.; Kumar, D.P.; Zhou, L.; Liu, Q. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J. Biol. Chem. 2012, 287, 5661–5672. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhu, H.; Sarbeng, E.B.; Liu, Q.; Tian, X.; Yang, Y.; Lyons, C.; Zhou, L.; Liu, Q. An unexpected second binding site for polypeptide substrates is essential for Hsp70 chaperone activity. J. Biol. Chem. 2020, 295, 584–596. [Google Scholar] [CrossRef]
- Mabate, B.; Zininga, T.; Ramatsui, L.; Makumire, S.; Achilonu, I.; Dirr, H.W.; Shonhai, A. Structural and biochemical charac-terization of Plasmodium falciparum Hsp70-x reveals functional versatility of its C-terminal EEVN motif. Proteins 2018, 86, 1189–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goeckeler, J.L.; Petruso, A.P.; Aguirre, J.; Clement, C.C.; Chiosis, G.; Brodsky, J.L. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 2008, 582, 2393–2396. [Google Scholar] [CrossRef] [Green Version]
- Dragovic, Z.; Broadley, S.A.; Shomura, Y.; Bracher, A.; Hartl, F.U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006, 25, 2519–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasco, L.; Dublang, L.; Moro, F.; Muga, A. The complex phosphorylation patterns that regulate the activity of Hsp70 and its cochaperones. Int. J. Mol. Sci. 2019, 20, 4122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattoo, R.U.; Farina Henriquez Cuendet, A.; Subanna, S.; Finka, A.; Priya, S.; Sharma, S.K.; Goloubinoff, P. Synergism between a foldase and an unfoldase: Reciprocal dependence between the thioredoxin-like activity of DnaJ and the polypeptide-unfolding activity of DnaK. Front. Mol. Biosci. 2014, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Kummer, E.; Bukau, B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015, 2, 22. [Google Scholar] [CrossRef] [Green Version]
- Shonhai, A.; Boshoff, A.; Blatch, G.L. The structural and functional diversity of Hsp70 proteins from Plasmodium falciparum. Protein Sci. 2007, 16, 1803–1818. [Google Scholar] [CrossRef] [Green Version]
- Kampinga, H.H.; Bergink, S. Heat shock proteins as potential targets for protective strategies in neurodegeneration. Lancet Neurol. 2016, 15, 748–759. [Google Scholar] [CrossRef]
- Garcia, V.M.; Nillegoda, N.B.; Bukau, B.; Morano, K.A. Substrate binding by the yeast Hsp110 nucleotide exchange factor and molecular chaperone Sse1 is not obligate for its biological activities. Mol. Biol. Cell 2017, 28, 2066–2075. [Google Scholar] [CrossRef] [Green Version]
- Polier, S.; Dragovic, Z.; Hartl, F.U.; Bracher, A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 2008, 133, 1068–1079. [Google Scholar] [CrossRef] [Green Version]
- Moenner, M.; Pluquet, O.; Bouchecareilh, M.; Chevet, E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res 2007, 67, 10631–10634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakes, R.S.; Bushnell, G.G.; Orbach, S.M.; Kandagatla, P.; Zhang, Y.; Morris, A.H.; Hall, M.S.; Lafaire, P.; Decker, J.T.; Hartfield, R.M.; et al. Metastatic conditioning of myeloid cells at a subcutaneous synthetic niche reflects disease progression and predicts pherapeutic outcomes. Cancer Res. 2020, 80, 602–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viale, A.; Draetta, G.F. Sugar? No Thank You, just a deep breath of oxygen for cancer stem cells. Cell Metab. 2015, 22, 543–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zininga, T.; Achilonu, I.; Hoppe, H.C.; Prinsloo, E.; Dirr, H.W.; Shonhai, A. Overexpression, purification and characterisation of the Plasmodium falciparum Hsp70-z (PfHsp70-z) protein. PLoS ONE 2015, 10, e0129445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Peter, J.J.; Sagar, A.; Ray, A.; Jha, M.P.; Rebeaud, M.; Tiwari, S.; Goloubinoff, P.; Ashish, F.; Mapa, K. Interdomain communication suppressing high intrinsic ATPase activity of Sse1 is essential for its co-disaggregase activity with Ssa1. FEBS J. 2019, 287, 671–694. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, K.; Nonoguchi, K.; Higashitsuji, H.; Kaneko, Y.; Sakurai, T.; Sumitomo, Y.; Itoh, K.; Subjeck, J.R.; Fujita, J. Apg-2 has a chaperone-like activity similar to Hsp110 and is overexpressed in hepatocellular carcinomas. FEBS Lett. 2004, 560, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Ullmann, R.; Morbini, P.; Halbwedl, I.; Bongiovanni, M.; Gogg-Kammerer, M.; Papotti, M.; Gabor, S.; Renner, H.; Popper, H.H. Protein expression profiles in adenocarcinomas and squamous cell carcinomas of the lung generated using tissue microarrays. J. Pathol. 2004, 203, 798–807. [Google Scholar] [CrossRef]
- Muchemwa, F.C.; Nakatsura, T.; Fukushima, S.; Nishimura, Y.; Kageshita, T.; Ihn, H. Differential expression of heat shock protein 105 in melanoma and melanocytic naevi. Melanoma Res. 2008, 18, 166–171. [Google Scholar] [CrossRef]
- Chan, D.S.M.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and Processed Meat and Colorectal Cancer Incidence: Meta-Analysis of Prospective Studies. PLoS ONE 2011, 6, e20456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, A.; Ogata, K.; Altan, B.; Yokobori, T.; Ide, M.; Mochiki, E.; Toyomasu, Y.; Kogure, N.; Yanoma, T.; Suzuki, M.; et al. Nuclear heat shock protein 110 expression is associated with poor prognosis and chemotherapy resistance in gastric cancer. Oncotarget 2016, 7, 18415. [Google Scholar] [CrossRef] [Green Version]
- Berthenet, K.; Bokhari, A.; Lagrange, A.; Marcion, G.; Boudesco, C.; Causse, S.; De Thonel, A.; Svrcek, M.; Goloudina, A.R.; Dumont, S.; et al. HSP110 promotes colorectal cancer growth through STAT3 activation. Oncogene 2017, 36, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.T. Turning up the heat on colorectal cancer. Nat. Med. 2011, 17, 1186–1188. [Google Scholar] [CrossRef]
- Duval, A.; Collura, A.; Berthenet, K.; Lagrange, A.; Garrido, C. Microsatellite instability in colorectal cancer: Time to stop hiding! Oncotarget 2011, 2, 826–827. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Watanabe, Y.; Tsujimura, A.; Tanaka, M. Expression of α-synuclein is regulated in a neuronal cell type-dependent manner. Anat. Sci. Int. 2018, 94, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Xu, K. Alpha-synuclein contributes to malignant progression of human meningioma via the Akt/mTOR path-way. Cancer Cell Int. 2016, 16, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, N.; Kakunda, M.; Pham, V.; Lill, J.R.; Du, P.; Wongchenko, M.J.; Yan, Y.; Firestein, R.; Huang, X. HSP105 recruits protein phosphatase 2A to dephosphorylate β-catenin. Mol. Cell. Biol. 2015, 35, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Kai, M.; Nakatsura, T.; Egami, H.; Senju, S.; Nishimura, Y.; Ogawa, M. Heat shock protein 105 is overexpressed in a variety of human tumors. Oncol. Rep. 2003, 10, 1777–1782. [Google Scholar] [CrossRef]
- Skrzypczak, M.; Goryca, K.; Rubel, T.; Paziewska, A.; Mikula, M.; Jarosz, D.; Pachlewski, J.; Oledzki, J.; Ostrowsk, J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE 2010, 5, e13091. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.M.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Kamran, M.Z.; Patil, P.; Gude, R.P. Role of STAT3 in cancer metastasis and translational advances. BioMed Res. Int. 2013, 2013, 421821. [Google Scholar] [CrossRef]
- Lakkim, V.; Reddy, M.C.; Prasad, D.V.R.; Lomada, D. Role of STAT3 in colorectal cancer development. In Role of Transcription Factors in Gastrointestinal Malignancies; Springer Nature: Singapore, 2017; pp. 269–298. [Google Scholar]
- Ma, J.-H.; Qin, L.; Li, X. Role of STAT3 signaling pathway in breast cancer. Cell Commun. Signal. 2020, 18, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Zhang, F.; Niu, R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci. Rep. 2016, 5, 17663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Li, G.; Huang, C.; Hou, Z.; Yang, X.; Luo, X.; Feng, Y.; Wang, G.; Hu, J.-B.; Cao, Z. The autophagy-independent role of BECN1 in colorectal cancer metastasis through regulating STAT3 signaling pathway activation. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zuo, D.; Subjeck, J.; Wang, X.Y. Unfolding the role of large heat shock proteins: New insights and therapeutic implications. Front. Immunol. 2016, 7, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosaka, S.; Nakatsura, T.; Tsukamoto, H.; Hatayama, T.; Baba, H.; Nishimura, Y. Synthetic small interfering RNA targeting heat shock protein 105 induces apoptosis of various cancer cells both in vitro and in vivo. Cancer Sci. 2006, 97, 623–632. [Google Scholar] [CrossRef]
- Entschladen, F.; Drell, T.L.; Lang, K.; Joseph, J.; Zaenker, K.S. Tumor-cell migration, invasion, and metastasis: Navigation by neurotransmitters. Lancet Oncol. 2004, 5, 254–258. [Google Scholar] [CrossRef]
- Spano, D.; Heck, C.; De Antonellis, P.; Christofori, G.; Zollo, M. Molecular networks that regulate cancer metastasis. Semin. Cancer Biol. 2012, 22, 234–249. [Google Scholar] [CrossRef]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef]
- De Matteis, S.; Canale, M.; Verlicchi, A.; Bronte, G.; Delmonte, A.; Crinò, L.; Martinelli, G.; Ulivi, P. Advances in molecular mechanisms and immunotherapy involving the immune cell-promoted epithelial-to-mesenchymal transition inlung cancer. J. Oncol. 2019, 2019, 7475364. [Google Scholar] [CrossRef]
- Manjili, M.H.; Park, J.; Facciponte, J.G.; Subjeck, J.R. HSP110 induces “danger signals” upon interaction with antigen presenting cells and mouse mammary carcinoma. Immunobiology 2005, 210, 295–303. [Google Scholar] [CrossRef]
- Gao, P.; Sun, X.; Chen, X.; Subjeck, J.; Wang, X.-Y. Secretion of stress protein grp170 promotes immune-mediated inhibition of murine prostate tumor. Cancer Immunol. Immunother. 2009, 58, 1319–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, D.; Zhang, S.; Jing, Z.; Shang, L.; Jin, S.; Liu, F.; Shen, J.; Li, Y.; Hu, J.; Meng, Q.; et al. Macrophages induce EMT to promote invasion of lung cancer cells through the IL-6-mediated COX-2/PGE2/β-catenin signaling pathway. Mol. Immunol. 2017, 90, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Browning, L.; Patel, M.R.; Horvath, E.B.; Tawara, K.; Jorcyk, C.L. IL-6 and ovarian cancer: Inflammatory cytokines in promotion of metastasis. Cancer Manag. Res. 2018, 10, 6685–6693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Safa, A.R. Epithelial-mesenchymal transition: A hallmark in pancreatic cancer stem cell migration, metastasis formation, and drug resistance. J. Cancer Metastasis Treat. 2020. [Google Scholar] [CrossRef]
- Lee, T.K.; Poon, R.T.P.; Yuen, A.P.; Ling, M.T.; Kwok, W.K.; Wang, X.H.; Wong, Y.C.; Guan, X.Y.; Man, K.; Chau, K.L.; et al. Twist overexpression correlates with hepatocel-lular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin. Cancer Res. 2006, 12, 5369–5376. [Google Scholar] [CrossRef] [Green Version]
- Hugo, H.; Ackland, M.L.; Blick, T.; Lawrence, M.G.; Clements, J.A.; Williams, E.D.; Thompson, E.W. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J. Cell Physiol. 2007, 213, 374–383. [Google Scholar] [CrossRef]
- Duennwald, M.L.; Echeverria, A.; Shorter, J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012, 10, e1001346. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Shen, F.; Yin, Y.X.; Yang, Y.Y.; Xiang, D.J.; Chen, Q. Increased expression of heat shock protein 27 correlates with peritoneal metastasis in epithelial ovarian cancer. Reprod. Sci. 2012, 19, 748–753. [Google Scholar] [CrossRef]
- Lopez-Soto, A.; Gonzalez, S.; Smyth, M.J.; Galluzzi, L. Control of metastasis by NK Cells. Cancer Cell 2017, 32, 135–154. [Google Scholar] [CrossRef]
- Komuro, A.; Yashiro, M.; Iwata, C.; Morishita, Y.; Johansson, E.; Matsumoto, Y.; Watanabe, A.; Aburatani, H.; Miyoshi, H.; Kiyono, K.; et al. Diffuse-type gastric carcinoma: Progression, angiogenesis, and transforming growth factor β signaling. J. Natl. Cancer Inst. 2009, 101, 592–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Li, Z.; Li, N.; Li, Y.; Chang, A.; Zhao, T.; Wang, X.; Wang, H.; Gao, S.; Yang, S.; et al. Interleukin 35 expression correlates with microvessel density in pancreatic ductal adenocarcinoma, recruits monocytes, and promotes growth and angiogenesis of xenograft tumors in mice. Gastroenterology 2018, 154, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Borges, T.J.; Wieten, L.; Van Herwijnen, M.J.C.; Broere, F.; Van Der Zee, R.; Bonorino, C.; Van Eden, W. The anti-inflammatory mechanisms of Hsp70. Front. Immunol. 2012, 3, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatayama, T.; Yamagishi, N.; Minobe, E.; Sakai, K. Role of hsp105 in protection against stress-induced apoptosis in neuronal PC12 cells. Biochem. Biophys. Res. Commun. 2001, 288, 528–534. [Google Scholar] [CrossRef]
- Yamagishi, N.; Ishihara, K.; Saito, Y.; Hatayama, T. Hsp105 family proteins suppress staurosporine-induced apoptosis by inhibiting the translocation of Bax to mitochondria in HeLa cells. Exp. Cell Res. 2006, 312, 3215–3223. [Google Scholar] [CrossRef]
- Boudesco, C.; Verhoeyen, E.; Martin, L.; Chassagne-Clement, C.; Salmi, L.; Mhaidly, R.; Pangault, C.; Fest, T.; Ramla, S.; Jardin, F.; et al. HSP110 sustains chronic NF-κB signaling in activated B-cell diffuse large B-cell lymphoma through MyD88 stabilization. Blood 2018, 132, 510–520. [Google Scholar] [CrossRef]
- Park, J.; Easton, D.; Chen, X.; Macdonald, I.J.; Wang, X.-Y.; Subjeck, J.R. The Chaperoning properties of mouse Grp170, a member of the third family of Hsp70 related proteins. Biochemistry 2003, 42, 14893–14902. [Google Scholar] [CrossRef]
- Behnke, J.; Hendershot, L.M. The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s. J. Biol. Chem. 2014, 289, 2899–2907. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.S. Glucose-regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nat. Rev. Cancer 2014, 14, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Tsukamoto, Y.; Kuwabara, K.; Hirota, S.; Kawano, K.; Yoshikawa, K.; Ozawa, K.; Kobayashi, T.; Yanagi, H.; Stern, D.M.; Tohyama, M.; et al. Expression of the 150-kd oxygen-regulated protein in human breast cancer. Lab. Investig. 1998, 78, 699–706. [Google Scholar]
- Stojadinovic, A.; Hooke, J.A.; Shriver, C.D.; Nissan, A.; Kovatich, A.J.; Kao, T.C. HYOU1/Orp150 expression in breast cancer. Med. Sci. Monit. 2007, 13, BR231–BR239. [Google Scholar] [PubMed]
- Facciponte, J.G.; Wang, X.-Y.; Subjeck, J.R. Hsp110 and Grp170, members of the Hsp70 superfamily, bind to scavenger receptor-A and scavenger receptor expressed by endothelial cells-I. Eur. J. Immunol. 2007, 37, 2268–2279. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, K.; Kuwabara, K.; Tamatani, M.; Takatsuji, K.; Tsukamoto, Y.; Kaneda, S.; Yanagi, H.; Stern, D.M.; Eguchi, Y.; Tsujimoto, Y.; et al. 150-kDa oxygen-regulated protein (ORP150) suppresses hypoxia-induced apoptotic cell death. J. Biol. Chem. 1999, 274, 6397–6404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozawa, K.; Tsukamoto, Y.; Hori, O.; Kitao, Y.; Yanagi, H.; Stern, D.M.; Ogawa, S. Regulation of tumor angiogenesis by oxygen-regulated protein 150, an inducible endoplasmic reticulum chaperone. Cancer Res. 2001, 61, 4206–4213. [Google Scholar]
- Miyagi, T.; Hori, O.; Koshida, K.; Egawa, M.; Kato, H.; Kitagawa, Y.; Ozawa, K.; Ogawa, S.; Namiki, M. Antitumor effect of reduction of 150-kDa oxygen-regulated protein expression on human prostate cancer cells. Int. J. Urol. 2002, 9, 577–585. [Google Scholar] [CrossRef]
- Asahi, H.; Koshida, K.; Hori, O.; Ogawa, S.; Namiki, M. Immunohistochemical detection of the 150-kDa oxygen-regulated protein in bladder cancer. BJU Int. 2002, 90, 462–466. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Ray, M.R.; Wafa, L.A.; Cheng, H.; Snoek, R.; Fazli, L.; Gleave, M.; Rennie, P.S. Cyclin G-associated kinase: A novel androgen receptor-interacting transcriptional coactivator that is overexpressed in hormone refractory prostate cancer. Int. J. Cancer 2005, 118, 1108–1119. [Google Scholar] [CrossRef]
- Dolly, S.O.; Gurden, M.D.; Drosopoulos, K.; Clarke, P.; De Bono, J.; Kaye, S.; Workman, P.; Linardopoulos, S. RNAi screen reveals synthetic lethality between cyclin G-associated kinase and FBXW7 by inducing aberrant mitoses. Br. J. Cancer 2017, 117, 954–964. [Google Scholar] [CrossRef] [Green Version]
- Wente, S.R.; Rout, M.P. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb. Perspect. Biol. 2010, 2, a000562. [Google Scholar] [CrossRef]
- Köhler, A.; Hurt, E. Gene regulation by nucleoporins and links to cancer. Mol. Cell 2010, 38, 6–15. [Google Scholar] [CrossRef]
- Rodriguez-Bravo, V.; Pippa, R.; Song, W.-M.; Carceles-Cordon, M.; Dominguez-Andres, A.; Fujiwara, N.; Woo, J.; Koh, A.P.; Ertel, A.; Lokareddy, R.K.; et al. Nuclear pores promote lethal prostate cancer by increasing POM121-driven E2F1, MYC, and AR nuclear import. Cell 2018, 174, 1200–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Li, L.; Jia, L.; Gong, A.; Wang, A.; Zhang, L.; Gu, M.; Tang, G. POM121 is identified as a novel prognostic marker of oral squamous cell carcinoma. J. Cancer 2019, 10, 4473–4480. [Google Scholar] [CrossRef] [PubMed]
- Nofrini, V.; Di Giacomo, D.; Mecucci, C. Nucleoporin genes in human diseases. Eur. J. Hum. Genet 2016, 24, 1388–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.-J.; Feng, Y.-H.; Gu, B.-H.; Li, Y.-M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linxweiler, J.; Körbel, C.; Müller, A.; Jüngel, E.; Blaheta, R.; Heinzelmann, J.; Stöckle, M.; Junker, K.; Menger, M.D.; Saar, M. Experimental imaging in orthotopic renal cell carcinoma xenograft models: Comparative evaluation of high-resolution 3D ultrasonography, in-vivo micro-CT and 9.4T MRI. Sci. Rep. 2017, 7, 14249. [Google Scholar] [CrossRef] [Green Version]
- Diwadkar-Navsariwala, V.; Prins, G.S.; Swanson, S.M.; Birch, L.A.; Ray, V.H.; Hedayat, S.; Lantvit, D.L.; Diamond, A.M. Selenoprotein deficiency accelerates prostate carcinogenesis in a transgenic model. Proc. Natl. Acad. Sci. USA 2006, 103, 8179–8184. [Google Scholar] [CrossRef] [Green Version]
- Tomala, K.; Korona, R. Molecular chaperones and selection against mutations. Biol. Direct. 2008, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.; Wang, T.; Araujo, T.L.S.; Sharma, S.; Brodsky, J.L.; Chiosis, G. Adapting to stress—Chaperome networks in cancer. Nat. Rev. Cancer 2018, 18, 562–575. [Google Scholar] [CrossRef]
- Pillarsetty, N.; Jhaveri, K.; Taldone, T.; Caldas-Lopes, E.; Punzalan, B.; Joshi, S.; Bolaender, A.; Uddin, M.M.; Rodina, A.; Yan, P.; et al. Paradigms for precision medicine in epichaperome cancer therapy. Cancer Cell 2019, 36, 559–573. [Google Scholar] [CrossRef]
- Yan, P.; Patel, H.J.; Sharma, S.; Corben, A.; Wang, T.; Panchal, P.; Yang, C.; Sun, W.; Araujo, T.L.; Rodina, A.; et al. Molecular stressors engender protein connectivity dysfunction through aberrant N-glycosylation of a chaperone. Cell Rep. 2020, 31, 107840. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Sekhar, A.; Nagesh, J.; Kay, L.E. Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. eLife 2017, 14, e28030. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Exosomal vesicles enhance immunosuppression in chronic inflammation: Impact in cellular senescence and the aging process. Cell. Signal. 2020, 75, 109771. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Subjeck, J.R. High molecular weight stress proteins: Identification, cloning and utilisation in cancer immunotherapy. Int. J. Hyperth. 2013, 29, 364–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, F.; Xu, Y.; Mao, L.; Ou, R.; Ding, Z.; Zhang, X.; Tang, J.; Li, B.; Jia, Z.; Tian, Z.; et al. Heat shock protein 110 improves the anti-tumor effects of the cytotoxic T lymphocyte epitope E7 in mice. Cancer Biol. 2010, 9, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Tang, S.; Le, S.Y.; Lu, R.; Rader, J.S.; Meyers, C.; Zheng, Z.M. Aberrant expression of oncogenic and tumour-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 2008, 3, e2557. [Google Scholar]
- Manjili, M.H.; Wang, X.Y.; Chen, X.; Martin, T.; Repasky, E.A.; Henderson, R.; Subjeck, J.R. HSP110-HER2/neu chaperone complex vaccine induces protective immunity against spontaneous mammary tumors in HER-2/neu transgenic mice. J. Immunol. 2003, 171, 4054–4061. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Y.; Kazim, L.; Repasky, E.A.; Subjeck, J.R. Immunization with tumor-derived ER chaperone grp170 elicits tumor-specific CD8+ T-cell responses and reduces pulmonary metastatic disease. Int. J. Cancer 2003, 105, 226–231. [Google Scholar] [CrossRef]
- Gao, P.; Sun, X.; Chen, X.; Wang, Y.; Foster, B.A.; Subjeck, J.; Fisher, P.B.; Wang, X.Y. Secretable chaperone Grp170 enhances therapeutic activity of a novel tumor suppressor, mda-7/IL-24. Cancer Res. 2008, 68, 3890–3898. [Google Scholar] [CrossRef] [Green Version]
- Dorard, C.; De Thonel, A.; Collura, A.; Marisa, L.; Svrcek, M.; Lagrange, A.; Jego, G.; Wanherdrick, K.; Joly, A.L.; Buhard, O.; et al. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat. Med. 2011, 17, 1283–1289. [Google Scholar] [CrossRef]
- Lang, B.J.; Guerrero-Giménez, M.E.; Prince, T.L.; Ackerman, A.; Bonorino, C.; Calderwood, S.K. Heat Shock Proteins are essential components in transformation and tumor progression: Cancer cell intrinsic pathways and beyond. Int. J. Mol. Sci. 2019, 20, 4507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zininga, T.; Shonhai, A. Small molecule inhibitors targeting the heat shock protein system of human obligate protozoan parasites. Int. J. Mol. Sci. 2019, 20, 5930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gozzi, G.J.; Gonzalez, D.; Boudesco, C.; Dias, A.M.M.; Gotthard, G.; Uyanik, B.; Dondaine, L.; Marcion, G.; Hermetet, F.; Denis, C.; et al. Selecting the first chemical molecule inhibitor of HSP110 for colorectal cancer therapy. Cell Death Differ. 2020, 27, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Ramatsui, L.; Makhado, P.B.; Makumire, S.; Achilonu, I.; Hoppe, H.C.; Dirr, H.W.; Shonhai, A. (−)-Epigallocatechin-3-gallate inhibits the chaperone activity of Plasmodium falciparum Hsp70 chaperones and abrogates their association with functional partners. Molecules 2017, 22, 2139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, S.W.; Gastpar, R.; Andreesen, R.; Gross, C.; Ullrich, H.; Thonigs, G.; Pfister, K.; Multhoff, G. Treatment of colon and lung cancer patients with ex vivo heat shock protein 70-peptide-activated, autologous natural killer cells. Clin. Cancer Res. 2004, 10, 3699–3707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solit, D.B.; Ivy, S.P.; Kopil, C.; Sikorski, R.; Morris, M.J.; Slovin, S.F.; Kelly, W.K.; DeLaCruz, A.; Curley, T.; Heller, G.; et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. Clin. Cancer Res. 2007, 13, 1775–1782. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, L.E.L.; Dingemans, A.M.C. Heat shock protein antagonists in early stage clinical trials for NSCLC. Expert. Opin. Investig. Drugs 2017, 26, 541–550. [Google Scholar] [CrossRef]
- Stangl, S.; Gehrmann, M.; Riegger, J.; Kuhs, K.; Riederer, I.; Sievert, W.; Hube, K.; Mocikat, R.; Dressel, R.; Kremmer, E. Targeting membrane heat-shock protein 70 (Hsp70) on tumors by cmHsp70.1 antibody. Proc. Natl. Acad. Sci. USA 2011, 108, 733–738. [Google Scholar] [CrossRef] [Green Version]



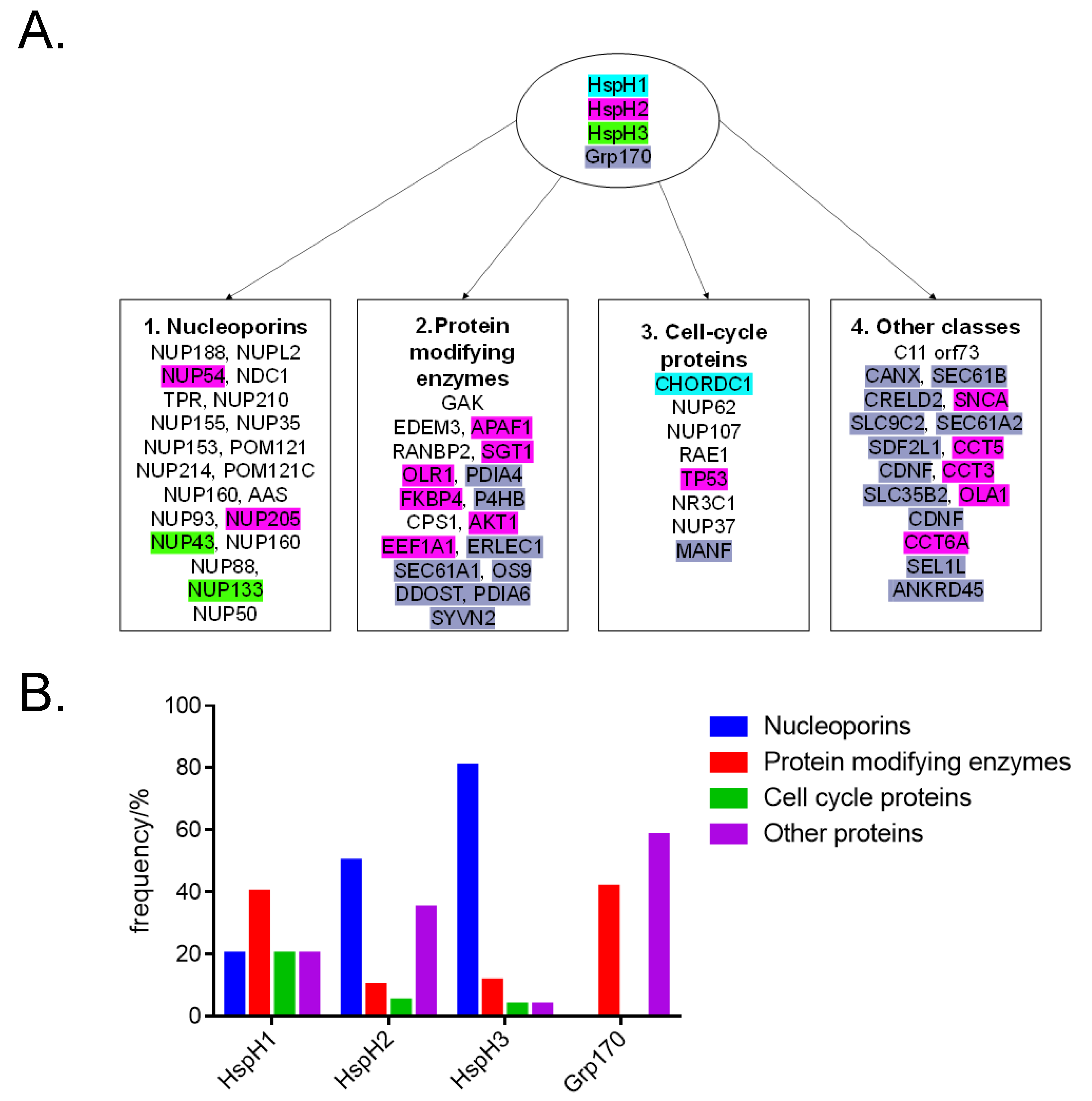
| Protein (Accession Number) | Size (kDa) | Localization | Stress Inducible (Yes/No) | Cellular Functions | References |
|---|---|---|---|---|---|
| 1. HspH1 (Q92598) | 97 | Cytosol, nucleus, endocytic vesicle | Yes | Apoptosis suppression, aggregation suppression, nucleotide exchange factor (NEF) | [46] |
| 2. HspH3 (O95757) | 95 | Cytosol, nucleus | Yes | Ellicits humoral immune responses in leukemia patients | [47] |
| 3. HspH2 (P34932) | 95 | Cytosol, extracellular exosome | ND | Implicated in spermatogenesis | [48] |
| 4. Grp170 (Q9Y4L1) | 111 | ER | Yes | Aggregation suppression, NEF | [49] |
| Protein | Function | Score |
|---|---|---|
| HspH1 Interaction Partners | ||
| 1. GAK-cyclin G (kinase) | Associates with cyclin G and CDK5 and is involved in the uncoating of clathrin-coated vesicles by Hsc70. | 0.879 |
| 2. CPS1(Carbamoyl-phosphate synthase) | Involved in the urea cycle and plays an important role in removing excess ammonia from the cell. | 0.874 |
| 3. EDEM3 (ER degradation-enhancing alpha-mannosidase-like protein 3 | Accelerates ER-associated degradation (ERAD) of glycoproteins by proteasomes. | 0.757 |
| 4. CHORDC1 (Cysteine and histidine-rich domain-containing protein 1) | Regulates centrosome duplication, probably by inhibiting the kinase activity of ROCK2. Proposed to act as co-chaperone for HSP90. Prevents tumorigenesis. | 0.747 |
| HspH3 Interaction Partners | ||
| 1. NUP188 (Nucleoporin) | May function as a component of the nuclear pore complex (NPC). | 0.937 |
| 2. C11 orf73 | Acts as a specific nuclear import carrier for HSP70. | 0.931 |
| 3. NUP37 (Nucleoporin) | Component of the Nup107-160 subcomplex of the nuclear pore complex (NPC) required for normal kinetochore microtubule attachment, mitotic progression and chromosome segregation. | 0.926 |
| 4. RANBP2 (E3 SUMO-protein ligase) | Facilitates SUMO1 and SUMO2 conjugation, (Ran-GTP, karyopherin)-mediated protein import. Component of the nuclear export pathway. | 0.924 |
| 5. TPR (Nucleoprotein TPR) | Essential for normal nucleocytoplasmic transport of proteins and mRNAs, plays a role in the establishment of nuclear-peripheral chromatin compartmentalization in interphase, and in the mitotic spindle checkpoint signaling during mitosis. | 0.917 |
| 7. RAE1 (mRNA export factor) | Plays a role in mitotic bipolar spindle formation. May function in nucleocytoplasmic transport. | 0.908 |
| 8. NUP155 (Nuclear pore complex protein) | May be essential for embryogenesis. Nucleoporins may be involved both in binding and translocating proteins during nucleocytoplasmic transport. | 0.907 |
| 9. NUP153 (Nuclear pore complex protein) | Essential for normal nucleocytoplasmic transport of proteins and mRNAs. Involved in the quality control and retention of unspliced mRNAs in the nucleus. | 0.904 |
| 10. NUP214 (Nuclear pore complex protein) | May serve as a docking site in the receptor-mediated import of substrates across the nuclear pore complex. | 0.904 |
| 11. NUP62 (Nuclear pore glycoprotein) | Plays a role in mitotic cell cycle progression by regulating centrosome segregation, centriole maturation and spindle orientation. It might be involved in protein recruitment to the centrosome after nuclear breakdown. | 0.904 |
| 12. NUP93 (Nuclear pore complex protein) | During renal development, regulates podocyte migration and proliferation through SMAD4 signaling. | 0.904 |
| 13. NUP43 (Nucleoporin) | Component of the Nup107-160 subcomplex of the nuclear pore complex (NPC) required for normal kinetochore microtubule attachment, mitotic progression and chromosome segregation. | 0.903 |
| 14. NUP88 (Nuclear pore complex protein) | Essential component of nuclear pore complex. | 0.903 |
| 15. NUP133 (Nuclear pore complex protein) | Involved in poly(A)+ RNA transport. | 0.903 |
| 16. NUP50 (Nuclear pore complex protein) | Interacts with regulatory proteins of cell cycle progression including CDKN1B. | 0.902 |
| 17. NUP107 (Nuclear pore complex protein) | Required for the assembly of peripheral proteins into the NPC. | 0.902 |
| 18. NDC1 (Nucleoporin) | Plays a key role in de novo assembly and insertion of NPC in the nuclear envelope. Required for NPC and nuclear envelope assembly, possibly by forming a link between the nuclear envelope membrane and soluble nucleoporins, thereby anchoring the NPC in the membrane. | 0.902 |
| 19. NUP210 (Nuclear pore membrane glycoprotein) | Essential for nuclear pore assembly and fusion, as well as structural integrity. | 0.901 |
| 20. NUP35 (Nucleoporin) | Can play the role of both NPC structural components and of docking or interaction partners for transiently associated nuclear transport factors. | 0.901 |
| 21. POM121 (Nuclear envelope pore membrane protein) | Essential component of the nuclear pore complex (NPC). May be involved in anchoring components of the pore complex to the pore membrane. | 0.900 |
| 22. POM121C (Nuclear envelope pore membrane protein) | Essential component of the nuclear pore complex (NPC). May be involved in anchoring components of the pore complex to the pore membrane. | 0.900 |
| 23. NUP160 (Nucleoporins) | Involved in poly(A)+ RNA transport. | 0.900 |
| 24. NUPL2 (Nucleoporin-like protein) | Required for the export of mRNAs containing poly(A) tails from the nucleus into the cytoplasm. | 0.900 |
| 25. AAS (Nucleoporin) | Plays a role in the normal development of the peripheral and central nervous system. | 0.900 |
| 26. GAK (Cyclin-G-associated kinase) | Involved in the uncoating of clathrin-coated vesicles by Hsc70 in non-neuronal cells. | 0.874 |
| 27. STIP1 (Stress-induced-phosphoprotein) | Mediates the association of the molecular chaperones HSPA8/HSC70 and HSP90. | 0857 |
| 28. EDEM3 (ER degradation-enhancing alpha-mannosidase-like protein 3) | Involved in endoplasmic reticulum-associated degradation (ERAD) of glycoproteins by proteasomes, by catalyzing mannose. | 0.792 |
| HspH2 Interaction Partners | ||
| 1. SNCA (Alpha-synuclein) | Induces fibrillization of microtubule-associated protein tau. Reduces neuronal responsiveness to various apoptotic stimuli, leading to a decreased caspase 3 activation. | 0.965 |
| 2. C11 orf73 | Acts as a specific nuclear import carrier for HSP70. | 0.964 |
| 3. NUP62 (Nuclear pore glycoprotein) | Plays a role in mitotic cell cycle progression by regulating centrosome segregation, centriole maturation and spindle orientation. It might be involved in protein recruitment to the centrosome after nuclear breakdown. | 0.944 |
| 4. RANBP2 (E3 SUMO-protein ligase) | Facilitates SUMO1 and SUMO2 conjugation, transport factor (Ran-GTP, karyopherin)-mediated protein import via the F-G repeat-containing domain which acts as a docking site for substrates. Component of the nuclear export pathway. | 0.940 |
| 5. TPR (Nucleoprotein) | Essential for normal nucleocytoplasmic transport of proteins and mRNAs, plays a role in the establishment of nuclear-peripheral chromatin compartmentalization in interphase, and in the mitotic spindle checkpoint signaling during mitosis. | 0.935 |
| 6. NUP37 (Nucleoporin) | Component of the Nup107-160 subcomplex of the nuclear pore complex (NPC) required for normal kinetochore microtubule attachment, mitotic progression and chromosome segregation. | 0.929 |
| 7. OLR1 (Oxidized low-density lipoprotein receptor) | Mediates the recognition, internalization and degradation of oxidatively modified low-density lipoprotein (oxLDL) by vascular endothelial cells. | 0.927 |
| 8. NUP155 (Nuclear pore complex protein) | Essential for embryogenesis. Nucleoporins may be involved both in binding and translocating proteins during nucleocytoplasmic transport. | 0.925 |
| 9. NUP54 (Nucleoporin p54) | Component of the nuclear pore complex, a complex required for the trafficking across the nuclear membrane. | 0.921 |
| 10. CCT2 (T-complex protein 1 subunit beta) | Molecular chaperone; assists the folding of proteins upon ATP hydrolysis. Known to play a role, in vitro, in the folding of actin and tubulin. | 0.917 |
| 11. RAE1 (mRNA export factor) | Plays a role in mitotic bipolar spindle formation. Binds mRNA. May function in nucleocytoplasmic transport. | 0.916 |
| 12. NUP107 (Nuclear pore complex protein) | Required for the assembly of peripheral proteins into the NPC. May anchor NUP62 to the NPC. | 0.916 |
| 13. NUP214 (Nuclear pore complex protein) | May serve as a docking site in the receptor-mediated import of substrates across the nuclear pore complex. | 0.915 |
| 14. NUP88 (Nucleoporins) | Essential component of nuclear pore complex. | 0.915 |
| 15. NUP93 (Nuclear pore complex protein) | During renal development, regulates podocyte migration and proliferation through SMAD4 signaling. | 0.914 |
| 16. AHSA1 (Activator of Hsp90 ATPase) | Activates the ATPase activity of HSP90AA1 leading to increase in its chaperone activity. | 0.913 |
| 17. NUP153 (Nuclear pore complex protein) | Essential for normal nucleocytoplasmic transport of proteins and mRNAs. Involved in the quality control and retention of unspliced mRNAs in the nucleus. | 0.912 |
| 18. NDC1 (Nucleoporin) | Plays a key role in de novo assembly and insertion of NPC in the nuclear envelope. | 0.907 |
| 19. NUP205 (Nuclear pore complex protein) | Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. | 0.907 |
| 20. NUP160 (Nuclear pore complex protein) | Involved in poly(A)+ RNA transport. | 0.907 |
| 21. NUP50 (Nuclear pore complex protein) | Interacts with regulatory proteins of cell cycle progression. | 0.906 |
| 22. NUP35 (Nucleoporin) | Can play the role of both NPC structural components and of docking or interaction partners for transiently associated nuclear transport factors. | 0.903 |
| 23. AAAS (Nucleoporins) | Plays a role in the normal development of the peripheral and central nervous system. | 0.903 |
| 24. FKBP4 (Peptidyl-prolyl cis-trans isomerase) | Immunophilin protein with PPIase. Plays a role in the intracellular trafficking of heterooligomeric forms of steroid hormone receptors between cytoplasm and nuclear compartments. Acts also as a regulator of microtubule dynamics by inhibiting MAPT/TAU ability to promote microtubule assembly. | 0.903 |
| 25. NUPL2 (Nucleoporin-like protein) | Required for the export of mRNAs containing poly(A) tails from the nucleus into the cytoplasm. | 0.903 |
| 26. NUP85 (Nuclear pore complex protein) | Required for spindle assembly during mitosis. | 0.903 |
| 27. NUP188 (Nucleoporin) | May function as a component of the NPC. | 0.903 |
| 28. NUP210 (Nuclear pore membrane glycoprotein) | Nucleoporin essential for nuclear pore assembly and fusion, nuclear pore spacing, as well as structural integrity. | 0.902 |
| 29. POM121C (Nuclear envelope pore membrane protein) | Essential component of the nuclear pore complex (NPC). | 0.902 |
| 30. POM121 (Nuclear envelope pore membrane protein) | Essential component of the nuclear pore complex (NPC). | 0.902 |
| 31. GAK (Cyclin-G-associated kinase) | Involved in the uncoating of clathrin-coated vesicles by Hsc70 in non-neuronal cells. | 0.893 |
| 32. CPS1 (Carbamoyl-phosphate synthase) | Involved in the urea cycle; plays an important role in removing excess ammonia from the cell. | 0.891 |
| 33. CLPB (Caseinolytic peptidase B protein) | May function as a regulatory ATPase and be related to secretion/protein trafficking process. | 0.889 |
| 34. AKT1 (RAC-alpha serine/threonine-protein kinase) | Regulates many processes including metabolism, proliferation, cell survival, growth and angiogenesis. AKT is responsible of the regulation of glucose uptake. | 0.883 |
| 35. CCT5 (T-complex protein 1 subunit epsilon) | Known to play a role, in vitro, in the folding of actin and tubulin. | 0.881 |
| 36. TP53 (Cellular tumor antigen p53) | Acts as a tumor suppressor in many tumor types; induces growth arrest or apoptosis depending on the physiological circumstances and cell type. Involved in cell cycle regulation as a trans-activator that acts to negatively regulate cell division by controlling a set of genes required for this process. | 0.881 |
| 37. CCT3 (T-complex protein 1 subunit gamma) | Known to play a role, in vitro, in the folding of actin and tubulin. | 0.856 |
| 38. EEF1A1 (Elongation factor 1-alpha 1) | Promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis. Forms a complex that acts as a T helper 1 (Th1) cell-specific transcription factor and binds the promoter of IFN-gamma to directly regulate its transcription and is thus involved importantly in Th1 cytokine production. | 0.853 |
| 39. CCT4 (T-complex protein 1 subunit delta) | Known to play a role, in vitro, in the folding of actin and tubulin. | 0.851 |
| 40. CCT6A (T-complex protein 1 subunit zeta) | Known to play a role, in vitro, in the folding of actin and tubulin. | 0.848 |
| 41. NR3C1 (Glucocorticoid receptor) | Has transcriptional repression activity. | 0.845 |
| 42. APAF1 (Apoptotic protease-activating factor 1) | Mediates the cytochrome c-dependent autocatalytic activation of procaspase 9 (Apaf-3), leading to the activation of caspase 3 and apoptosis. | 0.841 |
| 43. CFTR (Cystic fibrosis transmembrane conductance regulator) | Regulation of epithelial ion and water transport and fluid homeostasis. Mediates the transport of chloride ions across the cell membrane. | 0.836 |
| 44. SGT1 | May play a role in ubiquitination and subsequent proteasomal degradation of target proteins. | 0.820 |
| 45. OLA1 (Obg-like ATPase) | Hydrolyzes ATP, and can also hydrolyze GTP with lower efficiency. | 0.820 |
| Grp170 Interaction Partners | ||
| 1. PDIA4 (Protein disulphide-isomerase) | Belongs to the protein disulphide isomerase family. | 0.993 |
| 2. SIL1 (Nucleotide exchange factor) | Functions as a nucleotide exchange factor for the ER lumenal chaperone HSPA5. | 0.989 |
| 3. SEC63 (Translocation protein) | Required for integral membrane and secreted preprotein translocation across the endoplasmic reticulum membrane. | 0.962 |
| 4. P4HB (Protein disulphide-isomerase) | May cause structural modifications of exofacial proteins. Inside the cell, seems to form/rearrange disulphide bonds of nascent proteins. | 0.954 |
| 5. CALR (Calcium-binding chaperone) | Promotes folding, oligomeric assembly and quality control in the endoplasmic reticulum (ER) via the calreticulin/calnexin cycle. Interacts transiently with almost all of the monoglucosylated glycoproteins that are synthesized in the ER. | 0.954 |
| 6. PDIA6 (Protein disulphide-isomerase A6) | Negatively regulates the unfolded protein response (UPR) through binding to UPR sensors such as ERN1, which in turn inactivates ERN1 signaling. | 0.950 |
| 7. MANF (Mesencephalic astrocyte-derived neurotrophic factor) | Inhibits cell proliferation and endoplasmic reticulum (ER) stress-induced cell death (182 aa). | 0.946 |
| 8. CANX (Calcium-binding protein) | May act in assisting protein assembly and/or in the retention within the ER of unassembled protein subunits. It seems to play a major role in the quality control apparatus of the ER by the retention of incorrectly folded proteins. | 0.934 |
| 9. PDIA3 (Protein disulphide-isomerase A3) | Belongs to the protein disulphide isomerase family. | 0.924 |
| 10. SEC61A1 (Protein transport protein) | Plays a crucial role in the insertion of secretory and membrane polypeptides into the ER. Required for assembly of membrane and secretory proteins. | 0.906 |
| 11. CRELD2 | Cysteine rich with EGF-like domains | 0.901 |
| 12. SLC9C2 | Sodium/hydrogen exchange; Involved in pH regulation. | 0.876 |
| 13. ANKRD45 | Ankyrin repeat domain-containing protein. | 0.876 |
| 14. ERLEC1 (Endoplasmic reticulum lectin 1) | May function in endoplasmic reticulum quality control and endoplasmic reticulum-associated degradation (ERAD) of both non-glycosylated proteins and glycoproteins. | 0.876 |
| 15. CLGN (Calmegin) | Functions during spermatogenesis as a chaperone for a range of client proteins that. are important for sperm adhesion onto the egg zona pellucida and for subsequent penetration of the zona pellucida. | 0.868 |
| 16. DDOST (Dolichyl-diphosphooligosaccharide-protein glycosyltransferase) | Essential subunit of the N-oligosaccharyl transferase (OST) complex which catalyzes the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to an asparagine residue. Required for efficient N-glycosylation. | 0.866 |
| 17. SEC61A2 (Protein transport protein) | Plays a crucial role in the insertion of secretory and membrane polypeptides into the ER. It is required for assembly of membrane and secretory proteins. | 0.826 |
| 18. OS9 | Functions in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). | 0.803 |
| 19. SYVN2 (E3 ubiquitin-protein ligase synoviolin) | Component of the endoplasmic reticulum quality control (ERQC) system. Protects cells from ER stress-induced apoptosis. | 0.786 |
| 20. SDF2L1 | Stromal cell derived factor 2 like 1. | 0.783 |
| 21. SEL1L | Plays a role in the endoplasmic reticulum quality control (ERQC) system. Plays a role in LPL maturation and secretion. Required for normal differentiation and survival of pancreatic cells. | 0.783 |
| 22. CDNF (Cerebral dopamine neurotrophic factor) | Prevents the 6- hydroxydopamine (6-OHDA)-induced degeneration of dopaminergic neurons. Also prevents the degeneration of dopaminergic neurons. | 0.777 |
| 23. EDEM3 (ER degradation-enhancing alpha-mannosidase-like protein 3) | Involved in endoplasmic reticulum-associated degradation (ERAD). Accelerates the glycoprotein ERAD by proteasomes. | 0.773 |
| 24. SLC35B2 | May indirectly participate in activation of the NF- kappa-B and MAPK pathways. | 0.768 |
| 25. GAK (Cyclin-G-associated kinase) | Is involved in the uncoating of clathrin-coated vesicles by Hsc70 in non-neuronal cells. | 0.752 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakafana, G.; Shonhai, A. The Role of Non-Canonical Hsp70s (Hsp110/Grp170) in Cancer. Cells 2021, 10, 254. https://doi.org/10.3390/cells10020254
Chakafana G, Shonhai A. The Role of Non-Canonical Hsp70s (Hsp110/Grp170) in Cancer. Cells. 2021; 10(2):254. https://doi.org/10.3390/cells10020254
Chicago/Turabian StyleChakafana, Graham, and Addmore Shonhai. 2021. "The Role of Non-Canonical Hsp70s (Hsp110/Grp170) in Cancer" Cells 10, no. 2: 254. https://doi.org/10.3390/cells10020254







