Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Substrates and Enzymes
2.2. Enzyme Characterization
2.3. PU-PE Films Production
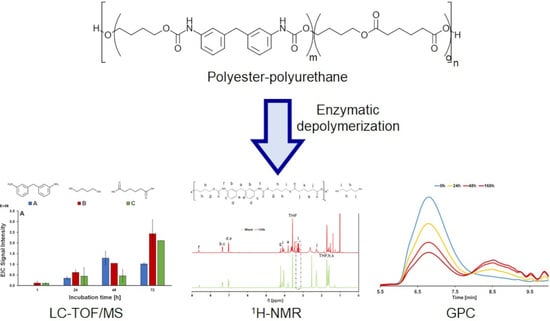
2.4. PU-PE Hydrolysis
2.5. Liquid Chromatography Time-of-Flight/Mass Spectrometry (LC-TOF/MS)
2.6. Gel Permeation Chromatography (GPC)
2.7. FT-IR Analysis
2.8. Scanning Electron Microscopy (SEM)
2.9. 1H-NMR Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soto, M.; Sebastián, R.M.; Marquet, J. Photochemical Activation of Extremely Weak Nucleophiles: Highly Fluorinated Urethanes and Polyurethanes from Polyfluoro Alcohols. J. Org. Chem. 2014, 79, 5019–5027. [Google Scholar] [CrossRef]
- Silva, A.L.; Bordado, J.C. Recent developments in polyurethane catalysis: Catalytic mechanisms review. Catal. Rev. 2004, 46, 31–51. [Google Scholar] [CrossRef]
- Seymour, R.B.; Kauffman, G.B. Polyurethanes: A class of modern versatile materials. J. Chem. Educ. 1992, 69, 909. [Google Scholar] [CrossRef]
- Eceiza, A.; de la Caba, K.; Kortaberria, G.; Gabilondo, N.; Marieta, C.; Corcuera, M.; Mondragon, I. Influence of molecular weight and chemical structure of soft segment in reaction kinetics of polycarbonate diols with 4,4’-diphenylmethane diisocyanate. Eur. Polym. J. 2005, 41, 3051–3059. [Google Scholar] [CrossRef]
- Polo, M.L.; Spontón, M.E.; Jaramillo, F.; Estenoz, D.A.; Meira, G.R. Linear segmented polyurethanes: I. A kinetics study. J. Appl. Polym. Sci. 2018, 135, 45747. [Google Scholar] [CrossRef]
- Shah, A.A.; Hasan, F.; Akhter, J.I.; Hameed, A.; Ahmed, S. Degradation of polyurethane by novel bacterial consortium isolated from soil. Ann. Microbiol. 2008, 58, 381–386. [Google Scholar] [CrossRef]
- Schmidt, J.; Wei, R.; Oeser, T.; e Silva, L.A.D.; Breite, D.; Schulze, A.; Zimmermann, W. Degradation of Polyester Polyurethane by Bacterial Polyester Hydrolases. Polymers 2017, 9, 65. [Google Scholar] [CrossRef] [Green Version]
- Akindoyo, J.O.; Beg, M.D.H.; Ghazali, S.; Islam, M.R.; Jeyaratnam, N.; Yuvaraj, A.R. Polyurethane types, synthesis and applications—A review. RSC Adv. 2016, 6, 114453–114482. [Google Scholar] [CrossRef] [Green Version]
- Buckley, C.; Prisacariu, C.; Martin, C. Elasticity and inelasticity of thermoplastic polyurethane elastomers: Sensitivity to chemical and physical structure. Polymer 2010, 51, 3213–3224. [Google Scholar] [CrossRef]
- Datta, J.; Kasprzyk, P. Thermoplastic polyurethanes derived from petrochemical or renewable resources: A comprehensive review. Polym. Eng. Sci. 2018, 58, E14–E35. [Google Scholar] [CrossRef] [Green Version]
- Gama, N.V.; Ferreira, A.; Barros-Timmons, A. Polyurethane foams: Past, present, and future. Materials 2018, 11, 1841. [Google Scholar] [CrossRef] [Green Version]
- Kausar, A. Polyurethane Composite Foams in High-Performance Applications: A Review. Polym. Technol. Eng. 2018, 57, 346–369. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Phalip, V.; Avérous, L. Evaluation of biological degradation of polyurethanes. Biotechnol. Adv. 2020, 39, 107457. [Google Scholar] [CrossRef] [PubMed]
- Datta, J.; Kopczyńska, P.; Simón, D.; Rodriguez, J.F. Thermo-Chemical Decomposition Study of Polyurethane Elastomer Through Glycerolysis Route with Using Crude and Refined Glycerine as a Transesterification Agent. J. Polym. Environ. 2018, 26, 166–174. [Google Scholar] [CrossRef] [Green Version]
- PlascticsEurope. Plastics-the Facts 2020 an Analysis of European Plastics Production, Demand and Waste Data. 2019. Available online: https://plasticseurope.org/knowledge-hub/plastics-the-facts-2020/ (accessed on 1 December 2021).
- Simón, D.; Borreguero, A.M.; de Lucas, A.; Rodríguez, J. Recycling of polyurethanes from laboratory to industry, a journey towards the sustainability. Waste Manag. 2018, 76, 147–171. [Google Scholar] [CrossRef]
- Kemona, A.; Piotrowska, M. Polyurethane Recycling and Disposal: Methods and Prospects. Polymers 2020, 12, 1752. [Google Scholar] [CrossRef]
- Yang, W.-S.; Lee, J.-S.; Park, S.-W.; Kang, J.-J.; Alam, T.; Seo, Y.-C. Gasification applicability study of polyurethane solid refuse fuel fabricated from electric waste by measuring syngas and nitrogenous pollutant gases. J. Mater. Cycles Waste Manag. 2016, 18, 509–516. [Google Scholar] [CrossRef]
- Gerlock, J.L.; Braslaw, J.; Mahoney, L.R.; Ferris, F.C. Reaction of polyurethane foam with dry steam: Kinetics and mechanism of reactions. J. Polym. Sci. 1980, 18, 541–557. [Google Scholar] [CrossRef]
- Mitova, V.; Grancharov, G.; Molero, C.; Borreguero, A.M.; Troev, K.; Rodriguez, J.F. Chemical degradation of polymers (Polyurethanes, polycarbonate and polyamide) by esters of H-phosphonic and phosphoric acids. J. Macromol. Sci. Part A 2013, 50, 774–795. [Google Scholar] [CrossRef]
- Simón, D.; Borreguero, A.M.; de Lucas, A.; Molero, C.; Rodríguez, J.F. Novel polyol initiator from polyurethane recycling residue. J. Mater. Cycles Waste Manag. 2014, 16, 525–532. [Google Scholar] [CrossRef]
- Gamerith, C.; Acero, E.H.; Pellis, A.; Ortner, A.; Vielnascher, R.; Luschnig, D.; Zartl, B.; Haernvall, K.; Zitzenbacher, S.; Strohmeier, G.; et al. Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption. Polym. Degrad. Stab. 2016, 132, 69–77. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150. [Google Scholar] [CrossRef]
- Akutsu, Y.; Nakajima-Kambe, T.; Nomura, N. Purification and Properties of a Polyester Polyurethane-Degrading Enzyme from Comamonas acidovorans TB-35. Appl. Environ. Microbiol. 1998, 64, 62–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Żądło-Dobrowolska, A.; Szczygieł, M.; Koszelewski, D.; Paprocki, D.; Ostaszewski, R. Self-immolative versatile fluorogenic probes for screening of hydrolytic enzyme activity. Org. Biomol. Chem. 2016, 14, 9146–9150. [Google Scholar] [CrossRef] [PubMed]
- Phua, S.K.; Castillo, E.; Anderson, J.M.; Hiltner, A. Biodegradation of a polyurethane in vitro. J. Biomed. Mater. Res. 1987, 21, 231–246. [Google Scholar] [CrossRef]
- Labow, R.S.; Erfle, D.J.; Santerre, J.P. Elastase-induced hvdrolvsis of synthetic solid sub & rate & poly (ester-urea-urethane) and poly (ether-urea-urethane). Biomaterials 1996, 17, 2381–2388. [Google Scholar]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Gamerith, C.; Vastano, M.; Ghorbanpour, S.M.; Zitzenbacher, S.; Ribitsch, D.; Zumstein, M.T.; Sander, M.; Acero, E.H.; Pellis, A.; Guebitz, G.M. Enzymatic Degradation of Aromatic and Aliphatic Polyesters by P. pastoris Expressed Cutinase 1 from Thermobifida cellulosilytica. Front. Microbiol. 2017, 8, 938. [Google Scholar] [CrossRef]
- Gigli, M.; Quartinello, F.; Soccio, M.; Pellis, A.; Lotti, N.; Guebitz, G.M.; Licoccia, S.; Munari, A. Enzymatic hydrolysis of poly (1,4-butylene 2,5-thiophenedicarboxylate) (PBTF) and poly (1,4-butylene 2,5-furandicarboxylate) (PBF) films: A comparison of mechanisms. Environ. Int. 2019, 130, 104852. [Google Scholar] [CrossRef]
- Weinberger, S.; Canadell, J.; Quartinello, F.; Yeniad, B.; Arias, A.; Pellis, A.; Guebitz, G.M. Enzymatic Degradation of Poly (ethylene 2,5-furanoate) Powders and Amorphous Films. Catalysts 2017, 7, 318. [Google Scholar] [CrossRef] [Green Version]
- Quartinello, F.; Vajnhandl, S.; Valh, J.V.; Farmer, T.; Vončina, B.; Lobnik, A.; Acero, E.H.; Pellis, A.; Guebitz, G.M. Synergistic chemo-enzymatic hydrolysis of poly (ethylene terephthalate) from textile waste. Microb. Biotechnol. 2017, 10, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Carniel, A.; Valoni, E.; Nicomedes, J.; da Conceição Gomes, A.; de Castro, A.M. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process. Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]
- Hunsen, M.; Azim, A.; Mang, H.; Wallner, S.R.; Ronkvist, A.; Xie, A.W.; Gross, R.A. A Cutinase with Polyester Synthesis Activity. Macromolecules 2007, 40, 148–150. [Google Scholar] [CrossRef] [Green Version]
- Gross, R.A.; Ganesh, M.; Lu, W. Enzyme-catalysis breathes new life into polyester condensation polymerizations. Trends Biotechnol. 2010, 28, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase structure, function and biocatalytic applications. Electron. J. Biotechnol. 1998, 1, 160–173. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, H.; Gupta, R.; Tiwari, A. Communities of Microbial Enzymes Associated with Biodegradation of Plastics. J. Polym. Environ. 2013, 21, 575–579. [Google Scholar] [CrossRef]
- Agrawal, P.B.; Nierstrasz, V.A.; Bouwhuis, G.H.; Warmoeskerken, M.M.C.G. Cutinase and pectinase in cotton bioscouring: An innovative and fast bioscouring process. Biocatal. Biotransform. 2008, 26, 412–421. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.; Araújo, R.; Casal, M.; Gübitz, G.M.; Cavaco-Paulo, A. Influence of mechanical agitation on cutinases and protease activity towards polyamide substrates. Enzym. Microb. Technol. 2007, 40, 1678–1685. [Google Scholar] [CrossRef] [Green Version]
- Ferrario, V.; Pellis, A.; Cespugli, M.; Guebitz, G.M.; Gardossi, L. Nature inspired solutions for polymers: Will cutinase enzymes make polyesters and polyamides greener? Catalysts 2016, 6, 205. [Google Scholar] [CrossRef] [Green Version]
- Quartinello, F.; Kremser, K.; Vecchiato, S.; Schoen, H.; Vielnascher, R.; PLoSzczanski, L.; Pellis, A.; Guebitz, G.M. Increased Flame Retardancy of Enzymatic Functionalized PET and Nylon Fabrics via DNA Immobilization. Front. Chem. 2019, 7, 685. [Google Scholar] [CrossRef]
- Pegoraro, M.; Galbiati, A.; Ricca, G. 1H nuclear magnetic resonance study of polyurethane prepolymers from toluene diisocyanate and polypropylene glycol. J. Appl. Polym. Sci. 2002, 87, 347–357. [Google Scholar] [CrossRef]
- Saito, T.; Aizawa, Y.; Tajima, K.; Isono, T.; Satoh, T. Organophosphate-catalyzed bulk ring-opening polymerization as an environmentally benign route leading to block copolyesters, end-functionalized polyesters, and polyester-based polyurethane. Polym. Chem. 2015, 6, 4374–4384. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Bisceglie, F.; Quartinello, F.; Vielnascher, R.; Guebitz, G.M.; Pellis, A. Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism. Polymers 2022, 14, 411. https://doi.org/10.3390/polym14030411
Di Bisceglie F, Quartinello F, Vielnascher R, Guebitz GM, Pellis A. Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism. Polymers. 2022; 14(3):411. https://doi.org/10.3390/polym14030411
Chicago/Turabian StyleDi Bisceglie, Federico, Felice Quartinello, Robert Vielnascher, Georg M. Guebitz, and Alessandro Pellis. 2022. "Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism" Polymers 14, no. 3: 411. https://doi.org/10.3390/polym14030411