KnowVolution of the Polymer-Binding Peptide LCI for Improved Polypropylene Binding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Library Generation
2.2. Site-Directed Mutagenesis
2.3. Expression of EGFP-10xAla-TEV-LCI in 96-Well Microtiter Plates
2.4. Expression of EGFP-10xAla-TEV-LCI in Flasks for Purification
2.5. Screening EGFP-10xAla-TEV-LCI for Improved Binding to Polypropylene in the Presence of Surfactant
2.6. Computational Analysis
2.7. Characterization of Purified EGFP-10xAla-TEV-LCI-Binding to Polypropylene in the Presence of Surfactant
3. Results
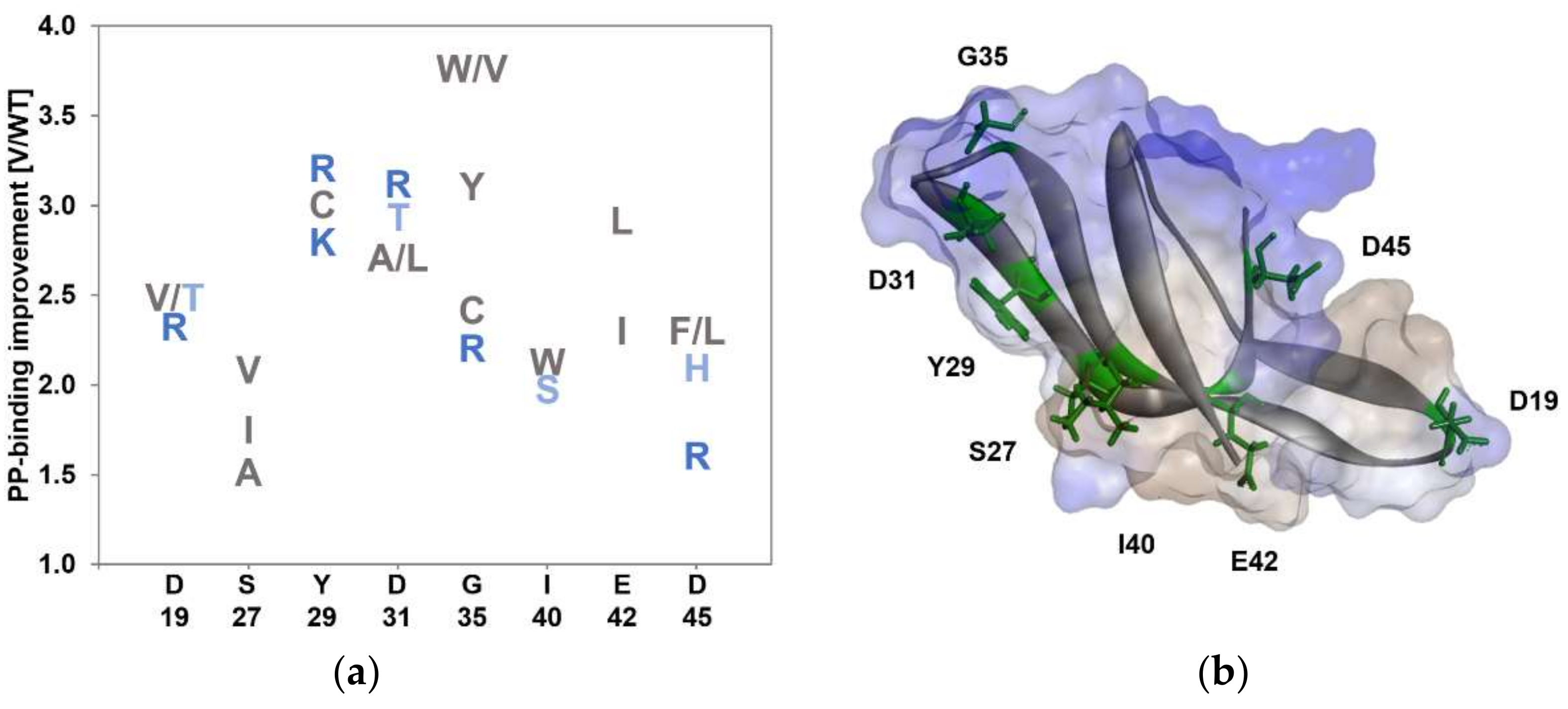
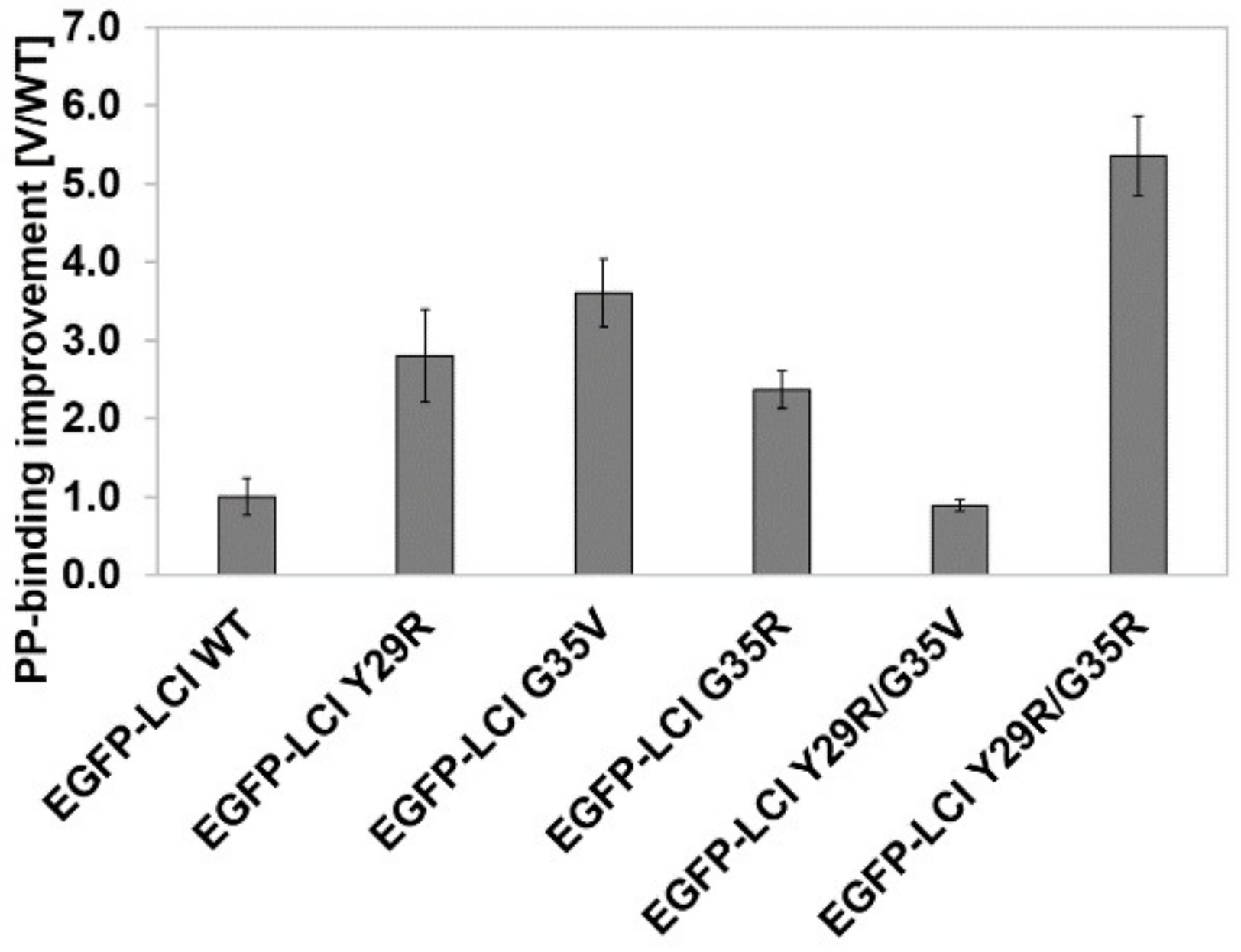
3.1. KnowVolution of the Anchor Peptide LCI for Improving Polypropylene-Binding Strength
3.2. Characterization of EGFP-10xAla-TEV-LCI and EGFP-10xAla-TEV-LCI KR-2-Binding to Polypropylene in the Presence of Triton X-100
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bolivar, J.M.; Gascon, V.; Marquez-Alvarez, C.; Blanco, R.M.; Nidetzky, B. Oriented Coimmobilization of Oxidase and Catalase on Tailor-Made Ordered Mesoporous Silica. Langmuir 2017, 33, 5065–5076. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Ma, S.Q.; Duan, S.; Deng, X.L.; Sun, Y.C.; Zhang, X.; Xu, X.H.; Guan, B.B.; Wang, C.; Hu, M.L.; et al. Modification of Titanium Substrates with Chimeric Peptides Comprising Antimicrobial and Titanium-Binding Motifs Connected by Linkers To Inhibit Biofilm Formation. ACS Appl. Mater. Interface 2016, 8, 5124–5136. [Google Scholar] [CrossRef] [PubMed]
- Care, A.; Bergquist, P.L.; Sunna, A. Solid-binding peptides: Smart tools for nanobiotechnology. Trends Biotechnol. 2015, 33, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Zernia, S.; Ott, F.; Bellmann-Sickert, K.; Frank, R.; Klenner, M.; Jahnke, H.G.; Prager, A.; Abel, B.; Robitzki, A.; Beck-Sickinger, A.G. Peptide-Mediated Specific Immobilization of Catalytically Active Cytochrome P450 BM3 Variant. Bioconjugate Chem. 2016, 27, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, A.B.; Miller, K.P.H.; Belcher, A.M.; Schmidt, C.E. Biomaterials functionalization using a novel peptide that selectively binds to a conducting polymer. Nat. Mater. 2005, 4, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Iijima, K.; Nagahama, H.; Takada, A.; Sawada, T.; Serizawa, T.; Hashizume, M. Surface functionalization of polymer substrates with hydroxyapatite using polymer-binding peptides. J. Mater. Chem. B 2016, 4, 3651–3659. [Google Scholar] [CrossRef]
- Serizawa, T.; Sawada, T.; Matsuno, H.; Matsubara, T.; Sato, T. A peptide motif recognizing a polymer stereoregularity. J. Am. Chem. Soc. 2005, 127, 13780–13781. [Google Scholar] [CrossRef] [PubMed]
- Serizawa, T.; Sawada, T.; Matsuno, H. Highly specific affinities of short peptides against synthetic polymers. Langmuir 2007, 23, 11127–11133. [Google Scholar] [CrossRef] [PubMed]
- Ejima, H.; Matsuno, H.; Serizawa, T. Biological Identification of Peptides that Specifically Bind to Poly(phenylene vinylene) Surfaces: Recognition of the Branched or Linear Structure of the Conjugated Polymer. Langmuir 2010, 26, 17278–17285. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Cui, Y. Recognition of epoxy with phage displayed peptides. Mat. Sci. Eng. C Mater. 2013, 33, 3082–3084. [Google Scholar] [CrossRef] [PubMed]
- Hnilova, M.; Oren, E.E.; Seker, U.O.S.; Wilson, B.R.; Collino, S.; Evans, J.S.; Tamerler, C.; Sarikaya, M. Effect of Molecular Conformations on the Adsorption Behavior of Gold-Binding Peptides. Langmuir 2008, 24, 12440–12445. [Google Scholar] [CrossRef] [PubMed]
- Menendez, A.; Scott, J.K. The nature of target-unrelated peptides recovered in the screening of phage-displayed random peptide libraries with antibodies. Anal. Biochem. 2005, 336, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Adey, N.B.; Mataragnon, A.H.; Rider, J.E.; Carter, J.M.; Kay, B.K. Characterization of Phage That Bind Plastic from Phage-Displayed Random Peptide Libraries. Gene 1995, 156, 27–31. [Google Scholar] [CrossRef]
- Serizawa, T.; Matsuno, H.; Sawada, T. Specific interfaces between synthetic polymers and biologically identified peptides. J. Mater. Chem. 2011, 21, 10252–10260. [Google Scholar] [CrossRef]
- Matsuno, H.; Sekine, J.; Yajima, H.; Serizawa, T. Biological selection of peptides for poly(L-lactide) substrates. Langmuir 2008, 24, 6399–6403. [Google Scholar] [CrossRef] [PubMed]
- Vodnik, M.; Strukelj, B.; Lunder, M. HWGMWSY, an unanticipated polystyrene binding peptide from random phage display libraries. Anal. Biochem. 2012, 424, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Rubsam, K.; Weber, L.; Jakob, F.; Schwaneberg, U. Directed evolution of polypropylene and polystyrene binding peptides. Biotechnol. Bioeng. 2018, 115, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, S.D.; Lowe, D.J.; O’Brien, J.P.; Wang, H.; Wilkins, A.E. Polypropylene Binding Peptides and Methods of Use. U.S. Patent 7928076B2, 19 April 2011. [Google Scholar]
- Rübsam, K.; Stomps, B.; Böker, A.; Jakob, F.; Schwaneberg, U. Anchor peptides: A green and versatile method for polypropylene functionalization. Polymer 2017, 116, 124–132. [Google Scholar] [CrossRef]
- Thai, C.K.; Dai, H.X.; Sastry, M.S.R.; Sarikaya, M.; Schwartz, D.T.; Baneyx, F. Identification and characterization of Cu2O- and ZnO-binding polypeptides by Escherichia coli cell surface display: Toward an understanding of metal oxide binding. Biotechnol. Bioeng. 2004, 87, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Baneyx, F.; Schwartz, D.T. Selection and analysis of solid-binding peptides. Curr. Opin. Biotech. 2007, 18, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Day, J.W.; Kim, C.H.; Smider, V.V.; Schultz, P.G. Identification of metal ion binding peptides containing unnatural amino acids by phage display. Bioorg. Med. Chem. Lett. 2013, 23, 2598–2600. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.B.; Wang, J.F.; Chen, Z.L.; Xia, B.; Lu, G.Y. Solution Structure of LCI, a Novel Antimicrobial Peptide from Bacillus subtilis. Biochemistry 2011, 50, 3621–3627. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Kishore, N. Thermodynamic insights into the binding of triton X-100 to globular proteins: A calorimetric and spectroscopic investigation. J. Phys. Chem. B 2006, 110, 9728–9737. [Google Scholar] [CrossRef] [PubMed]
- Mehan, S.; Aswal, V.K.; Kohlbrecher, J. Tuning of protein-surfactant interaction to modify the resultant structure. Phys. Rev. E 2015, 92, Artn-032713. [Google Scholar] [CrossRef] [PubMed]
- Horinek, D.; Serr, A.; Geisler, M.; Pirzer, T.; Slotta, U.; Lud, S.Q.; Garrido, J.A.; Scheibel, T.; Hugel, T.; Netz, R.R. Peptide adsorption on a hydrophobic surface results from an interplay of solvation, surface, and intrapeptide forces. Proc. Natl. Acad. Sci. USA 2008, 105, 2842–2847. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, E.A.; Mundhada, H.; Meier, T.; Duefel, H.; Bocola, M.; Schwaneberg, U. Reengineered glucose oxidase for amperometric glucose determination in diabetes analytics. Biosens. Bioelectron. 2013, 50, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Zhu, L.L.; Schwaneberg, U. Directed evolution 2.0: Improving and deciphering enzyme properties. Chem. Commun. 2015, 51, 9760–9772. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, C.; Sibilla, F.; Maugeri, Z.; Streit, W.R.; de Maria, P.D.; Martinez, R.; Schwaneberg, U. Reengineering CelA2 cellulase for hydrolysis in aqueous solutions of deep eutectic solvents and concentrated seawater. Green Chem. 2012, 14, 2719–2726. [Google Scholar] [CrossRef]
- Martinez, R.; Jakob, F.; Tu, R.; Siegert, P.; Maurer, K.H.; Schwaneberg, U. Increasing activity and thermal resistance of Bacillus gibsonii alkaline protease (BgAP) by directed evolution. Biotechnol. Bioeng. 2013, 110, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Malcolm, B.A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange (TM) site-directed mutagenesis. Biotechniques 1999, 26, 680–682. [Google Scholar] [PubMed]
- Cadwell, R.C.; Joyce, G.F. Mutagenic Pcr. PCR Meth. Appl. 1994, 3, S136–S140. [Google Scholar] [CrossRef]
- Kumada, Y.; Kuroki, D.; Yasui, H.; Ohse, T.; Kishimoto, M. Characterization of polystyrene-binding peptides (PS-tags) for site-specific immobilization of proteins. J. Biosci. Bioeng. 2010, 109, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Bakhshinejad, B.; Sadeghizadeh, M. A polystyrene binding target-unrelated peptide isolated in the screening of phage display library. Anal. Biochem. 2016, 512, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Choe, W.S.; Sastry, M.S.R.; Thai, C.K.; Dai, H.; Schwartz, D.T.; Baneyx, F. Conformational control of inorganic adhesion in a designer protein engineered for cuprous oxide binding. Langmuir 2007, 23, 11347–11350. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, M.B.; Jones, S.E.; Cai, Y.; Ahmad, G.; Naik, R.R.; Kroger, N.; Sandhage, K.H. Identification and design of peptides for the rapid, high-yield formation of nanoparticulate TiO2 from aqueous solutions at room temperature. Chem. Mater. 2008, 20, 1578–1584. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Nidetzky, B. Positively Charged Mini-Protein Z(basic2) As a Highly Efficient Silica Binding Module: Opportunities for Enzyme Immobilization on Unmodified Silica Supports. Langmuir 2012, 28, 10040–10049. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.W.; Ruaan, R.C.; Liu, C.I. Adsorption of Antimicrobial Indolicidin-Derived Peptides on Hydrophobic Surfaces. Langmuir 2012, 28, 10446–10452. [Google Scholar] [CrossRef] [PubMed]
- Dennig, A.; Shivange, A.V.; Marienhagen, J.; Schwaneberg, U. OmniChange: The sequence independent method for simultaneous site-saturation of five codons. PLoS ONE 2011, 6, e26222. [Google Scholar] [CrossRef] [PubMed]
- Kumada, Y.; Murata, S.; Ishikawa, Y.; Nakatsuka, K.; Kishimoto, M. Screening of PC and PMMA-binding peptides for site-specific immobilization of proteins. J. Biotechnol. 2012, 160, 222–228. [Google Scholar] [CrossRef] [PubMed]

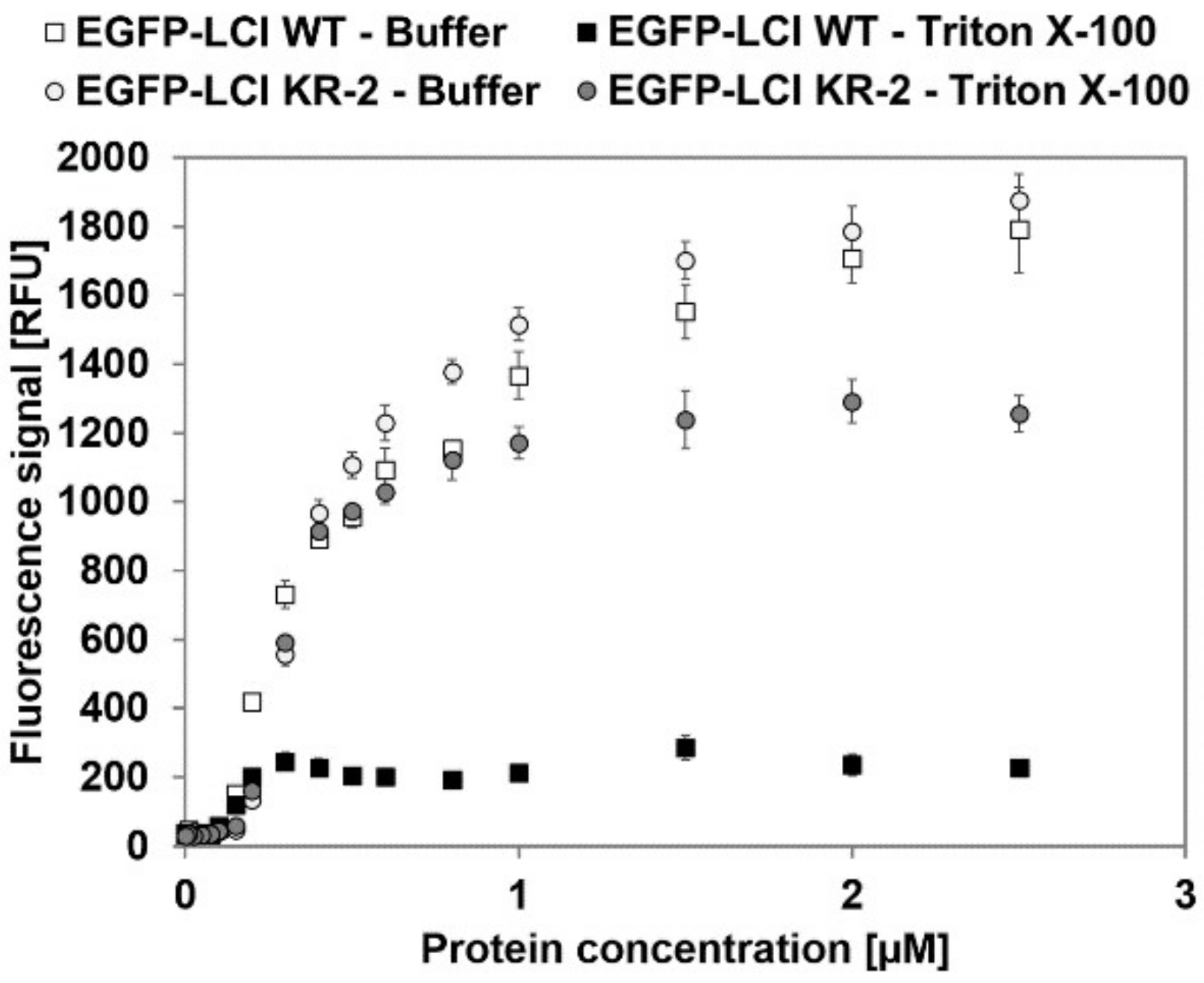
| Variant | Elution | BMax [pmol/cm2] | KD [µM] | R2 |
|---|---|---|---|---|
| EGFP-LCI WT | Buffer | 14.63 ± 0.30 | 0.47 ± 0.02 | 0.985 |
| Triton X-100 | 1.86 ± 0.06 | 0.18 ± 0.02 | 0.910 | |
| EGFP-LCI KR-2 | Buffer | 15.19± 0.56 | 0.54 ± 0.04 | 0.980 |
| Triton X-100 | 8.84 ± 0.05 | 0.34 ± 0.03 | 0.997 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rübsam, K.; Davari, M.D.; Jakob, F.; Schwaneberg, U. KnowVolution of the Polymer-Binding Peptide LCI for Improved Polypropylene Binding. Polymers 2018, 10, 423. https://doi.org/10.3390/polym10040423
Rübsam K, Davari MD, Jakob F, Schwaneberg U. KnowVolution of the Polymer-Binding Peptide LCI for Improved Polypropylene Binding. Polymers. 2018; 10(4):423. https://doi.org/10.3390/polym10040423
Chicago/Turabian StyleRübsam, Kristin, Mehdi D. Davari, Felix Jakob, and Ulrich Schwaneberg. 2018. "KnowVolution of the Polymer-Binding Peptide LCI for Improved Polypropylene Binding" Polymers 10, no. 4: 423. https://doi.org/10.3390/polym10040423





