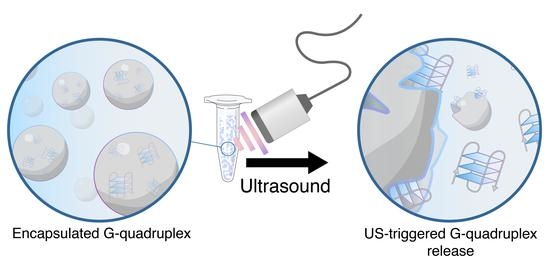
Encapsulation and Ultrasound-Triggered Release of G-Quadruplex DNA in Multilayer Hydrogel Microcapsules
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Double-stranded Calf Thymus DNA
2.3. Preparation of DNA Oligonucleotides
2.4. Encapsulation of DNA into Calcium Carbonate Microparticles
2.5. Synthesis of G-Quadruplex-Loaded (PMAA/PVPON)n Multilayer Hydrogel Capsules
2.6. Characterization of (PMAA/PVPON)13 Capsules Using Fourier Transform Infrared Spectroscopy (FTIR)
2.7. Characterization of (PMAA/PVPON)n Capsules by SEM and Confocal Laser Scanning Microscopy (CLSM)
2.8. ζ-potential Measurements of (PMAA/PVPON)n Multilayer Hydrogel Capsules
2.9. US-Controlled Release of G-Quadruplex DNA from the (PMAA/PVPON)n Capsules
2.10. Turbidimetry of GSH-Treated (PMAA/PVPON)13 Capsules
2.11. Quantification of G-Quadruplex DNA Release
2.12. Quantification of dsDNA Release
2.13. Probing Structural Changes of G-Quadruplex DNA after Release
3. Results and Discussion
3.1. Synthesis of (PMAA/PVPON)n Hydrogel Capsules Using Calcium Carbonate Sacrificial Templates
3.2. Encapsulation of Nucleotides into (PMAA/PVPON)n Hydrogel Capsules
3.3. Enzymatic Degradation of dsDNA- and G-Quadruplex-Loaded (PMAA/PVPON)n Hydrogel Capsules
3.4. Ultrasound (US)-Triggered Release of G-Quadruplex DNA from (PMAA/PVPON)n Hydrogel Capsules
3.5. Encapsulation and US-Triggered Release of dsDNA from (PMAA/PVPON)13 Capsules
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Breslauer, K.; Frank, R.; Blöcker, H.; Marky, L. Predicting DNA Duplex Stability from the Base Sequence. Proc. Natl. Acad. Sci. USA 1986, 83, 3746–3750. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; Zhang, S. Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003, 4, 566. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Willner, B.; Willner, I. DNA nanotechnology: From sensing and DNA machines to drug-delivery systems. ACS Nano 2013, 7, 8320–8332. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.P.; Actis, P.; Jönsson, P.; Klenerman, D.; Korchev, Y.; Edel, J.B. On-demand delivery of single DNA molecules using nanopipets. ACS Nano 2015, 9, 3587–3595. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.N.; Green, J.J.; Chan, J.M.; Langer, R.; Anderson, D.G. Polymeric materials for gene delivery and DNA vaccination. Adv. Mater. 2008, 21, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Bauhuber, S.; Hozsa, C.; Breunig, M.; Göpferich, A. Delivery of nucleic acids via disulfide-based carrier systems. Adv. Mater. 2009, 21, 3286–3306. [Google Scholar] [CrossRef] [PubMed]
- Mastorakos, P.; da Silva, A.L.; Chisholm, J.; Song, E.; Choi, W.K.; Boyle, M.P.; Morales, M.M.; Hanes, J.; Suk, J.S. Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 8720. [Google Scholar] [CrossRef] [PubMed]
- Alton, E.W.F.W.; Armstrong, D.K.; Ashby, D.; Bayfield, K.J.; Bilton, D.; Bloomfield, E.V.; Boyd, A.C.; Brand, J.; Buchan, R.; Calcedo, R.; et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: A randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015, 3, 684–691. [Google Scholar] [CrossRef]
- Han, H.; Hurley, L.H. G-quadruplex DNA: A potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000, 21, 136–142. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA g-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Maizels, N.; Gray, L.T. The g4 genome. PLOS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of g-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770. [Google Scholar] [CrossRef]
- Tucker, B.A.; Hudson, J.S.; Ding, L.; Lewis, E.; Sheardy, R.D.; Kharlampieva, E.; Graves, D. Stability of the na+ form of the human telomeric g-quadruplex: Role of adenines in stabilizing g-quadruplex structure. ACS Omega 2018, 3, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745. [Google Scholar] [CrossRef]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Düchler, M. G-quadruplexes: Targets and tools in anticancer drug design. J. Drug Target. 2012, 20, 389–400. [Google Scholar] [CrossRef]
- Graziella, C.-R.; Nadia, Z.; Marco, F. Emerging role of g-quadruplex DNA as target in anticancer therapy. Curr. Pharm. Des. 2016, 22, 6612–6624. [Google Scholar] [CrossRef]
- Buket, O.; Clement, L.I.N.; DanZhou, Y. DNA g-quadruplex and its potential as anticancer drug target. Sci. China Chem. 2014, 57, 1605–1614. [Google Scholar] [CrossRef]
- Herweijer, H.; Wolff, J.A. Progress and prospects: Naked DNA gene transfer and therapy. Gene Ther. 2003, 10, 453. [Google Scholar] [CrossRef]
- Wang, W.; Balk, M.; Deng, Z.; Wischke, C.; Gossen, M.; Behl, M.; Ma, N.; Lendlein, A. Engineering biodegradable micelles of polyethylenimine-based amphiphilic block copolymers for efficient DNA and sirna delivery. J. Control. Release 2016, 242, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Beavers Kelsey, R.; Werfel Thomas, A.; Shen, T.; Kavanaugh Taylor, E.; Kilchrist Kameron, V.; Mares Jeremy, W.; Fain Joshua, S.; Wiese Carrie, B.; Vickers Kasey, C.; Weiss Sharon, M.; et al. Porous silicon and polymer nanocomposites for delivery of peptide nucleic acids as anti-microrna therapies. Adv. Mater. 2016, 28, 7984–7992. [Google Scholar] [CrossRef] [PubMed]
- Lambricht, L.; Lopes, A.; Kos, S.; Sersa, G.; Préat, V.; Vandermeulen, G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expert Opin. Drug Deliv. 2016, 13, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Molla Mijanur, R.; Levkin Pavel, A. Combinatorial approach to nanoarchitectonics for nonviral delivery of nucleic acids. Adv. Mater. 2015, 28, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Lehto, T.; Ezzat, K.; Wood, M.J.A.; El Andaloussi, S. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev. 2016, 106, 172–182. [Google Scholar] [CrossRef]
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Hackett, P.B.; Largaespada, D.A.; Switzer, K.C.; Cooper, L.J.N. Evaluating risks of insertional mutagenesis by DNA transposons in gene therapy. Transl. Res. J. Lab. Clin. Med. 2013, 161, 265–283. [Google Scholar] [CrossRef] [Green Version]
- Hardee, C.L.; Arévalo-Soliz, L.M.; Hornstein, B.D.; Zechiedrich, L. Advances in non-viral DNA vectors for gene therapy. Genes 2017, 8, 65. [Google Scholar] [CrossRef]
- Keeney, M.; Ong, S.-G.; Padilla, A.; Yao, Z.; Goodman, S.; Wu, J.C.; Yang, F. Development of poly(β-amino ester)-based biodegradable nanoparticles for nonviral delivery of minicircle DNA. ACS Nano 2013, 7, 7241–7250. [Google Scholar] [CrossRef]
- Richardson Joseph, J.; Choy Mei, Y.; Guo, J.; Liang, K.; Alt, K.; Ping, Y.; Cui, J.; Law Lok, S.; Hagemeyer Christoph, E.; Caruso, F. Polymer capsules for plaque-targeted in vivo delivery. Adv. Mater. 2016, 28, 7703–7707. [Google Scholar] [CrossRef]
- Phillips, L.C.; Klibanov, A.L.; Wamhoff, B.R.; Hossack, J.A. Targeted gene transfection from microbubbles into vascular smooth muscle cells using focused, ultrasound-mediated delivery. Ultrasound Med. Boil. 2010, 36, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Xie, A.; Belcik, T.; Qi, Y.; Morgan, T.K.; Champaneri, S.A.; Taylor, S.; Davidson, B.P.; Zhao, Y.; Klibanov, A.L.; Kuliszewski, M.A.; et al. Ultrasound-mediated vascular gene transfection by cavitation of endothelial-targeted cationic microbubbles. JACC Cardiovasc. Imaging 2012, 5, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Kumon, R.E.; Deng, C.X. Mechanisms of microbubble-facilitated sonoporation for drug and gene delivery. Therap. Deliv. 2014, 5, 467–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlovskaya, V.; Kharlampieva, E.; Erel, I.; Sukhishvili, S.A. Multilayer-derived, ultrathin, stimuli-responsive hydrogels. Soft Matter 2009, 5, 4077–4087. [Google Scholar] [CrossRef]
- Kharlampieva, E.; Kozlovskaya, V.; Sukhishvili, S.A. Layer-by-layer hydrogen-bonded polymer films: From fundamentals to applications. Adv. Mater. 2009, 21, 3053–3065. [Google Scholar] [CrossRef]
- Lvov, Y.; Decher, G.; Sukhorukov, G. Assembly of thin films by means of successive deposition of alternate layers of DNA and poly(allylamine). Macromolecules 1993, 26, 5396–5399. [Google Scholar] [CrossRef]
- Lvov, Y.M.; Lu, Z.; Schenkman, J.B.; Zu, X.; Rusling, J.F. Direct electrochemistry of myoglobin and cytochrome p450cam in alternate layer-by-layer films with DNA and other polyions. J. Am. Chem. Soc. 1998, 120, 4073–4080. [Google Scholar] [CrossRef]
- Decher, G.; Lehr, B.; Lowack, K.; Lvov, Y.; Schmitt, J. New nanocomposite films for biosensors: Layer-by-layer adsorbed films of polyelectrolytes, proteins or DNA. Biosens. Bioelectron. 1994, 9, 677–684. [Google Scholar] [CrossRef]
- Lynn, D.M. Polyelectrolyte Multilayer Coatings for the Release and Transfer of Plasmid DNA. In Layer-by-Layer Films for Biomedical Applications; Picart, C., Caruso, F., Voegel, J., Eds.; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Appadoo, V.; Carter, M.C.D.; Lynn, D.M. Controlling the surface-mediated release of DNA using ‘mixed multilayers’. Bioeng. Transl. Med. 2016, 1, 181–192. [Google Scholar] [CrossRef]
- Yu, Y.; Si, Y.; Bechler, S.L.; Liu, B.; Lynn, D.M. Polymer multilayers that promote the rapid release and contact transfer of DNA. Biomacromolecules 2015, 16, 2998–3007. [Google Scholar] [CrossRef]
- Castleberry, S.A.; Golberg, A.; Sharkh, M.A.; Khan, S.; Almquist, B.D.; Austen, W.G.; Yarmush, M.L.; Hammond, P.T. Nanolayered sirna delivery platforms for local silencing of CTGF reduce cutaneous scar contraction in third-degree burns. Biomaterials 2016, 95, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.H.; Lee, J.B.; Shopsowitz, K.E.; Dreaden, E.C.; Morton, S.W.; Poon, Z.; Hong, J.; Yamin, I.; Bonner, D.K.; Hammond, P.T. Layer-by-layer assembled antisense DNA microsponge particles for efficient delivery of cancer therapeutics. ACS Nano 2014, 8, 9767–9780. [Google Scholar] [CrossRef] [PubMed]
- de Villiers, M.M.; Lvov, Y.M. Layer-by-layer self-assembled nanoshells for drug delivery. Adv. Drug Deliv. Rev. 2011, 63, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskaya, V.; Higgins, W.; Chen, J.; Kharlampieva, E. Shape switching of hollow layer-by-layer hydrogel microcontainers. Chem. Commun. 2011, 47, 8352–8354. [Google Scholar] [CrossRef]
- Chen, J.; Kozlovskaya, V.; Goins, A.; Campos-Gomez, J.; Saeed, M.; Kharlampieva, E. Biocompatible shaped particles from dried multilayer polymer capsules. Biomacromolecules 2013, 14, 3830–3841. [Google Scholar] [CrossRef]
- Vergaro, V.; Scarlino, F.; Bellomo, C.; Rinaldi, R.; Vergara, D.; Maffia, M.; Baldassarre, F.; Giannelli, G.; Zhang, X.; Lvov, Y.M.; et al. Drug-loaded polyelectrolyte microcapsules for sustained targeting of cancer cells. Adv. Drug Deliv. Rev. 2011, 63, 847–864. [Google Scholar] [CrossRef]
- Cavalieri, F.; Postma, A.; Lee, L.; Caruso, F. Assembly and functionalization of DNA−polymer microcapsules. ACS Nano 2009, 3, 234–240. [Google Scholar] [CrossRef]
- Kakade, S.; Manickam, D.S.; Handa, H.; Mao, G.; Oupický, D. Transfection activity of layer-by-layer plasmid DNA/poly(ethylenimine) films deposited on plga microparticles. Int. J. Pharm. 2009, 365, 44–52. [Google Scholar] [CrossRef]
- Elsner, N.; Kozlovskaya, V.; Sukhishvili, S.A.; Fery, A. pH-triggered softening of crosslinked hydrogen-bonded capsules. Soft Matter 2006, 2, 966–972. [Google Scholar] [CrossRef]
- Kozlovskaya, V.; Wang, Y.; Higgins, W.; Chen, J.; Chen, Y.; Kharlampieva, E. pH-triggered shape response of cubical ultrathin hydrogel capsules. Soft Matter 2012, 8, 9828–9839. [Google Scholar] [CrossRef]
- De Geest, B.G.; Skirtach, A.G.; Mamedov, A.A.; Antipov, A.A.; Kotov, N.A.; De Smedt, S.C.; Sukhorukov, G.B. Ultrasound-triggered release from multilayered capsules. Small 2007, 3, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskaya, V.; Shamaev, A.; Sukhishvili, S.A. Tuning swelling ph and permeability of hydrogel multilayer capsules. Soft Matter 2008, 4, 1499–1507. [Google Scholar] [CrossRef]
- Shchukin, D.G.; Patel, A.A.; Sukhorukov, G.B.; Lvov, Y.M. Nanoassembly of biodegradable microcapsules for DNA encasing. J. Am. Chem. Soc. 2004, 126, 3374–3375. [Google Scholar] [CrossRef] [PubMed]
- Zelikin, A.N.; Li, Q.; Caruso, F. Degradable polyelectrolyte capsules filled with oligonucleotide sequences. Angew. Chem. Int. Ed. 2006, 45, 7743–7745. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.L.; Such, G.K.; Johnston, A.P.R.; Antequera-García, G.; Caruso, F. Controlled release of DNA from poly(vinylpyrrolidone) capsules using cleavable linkers. Biomaterials 2011, 32, 6277–6284. [Google Scholar] [CrossRef]
- Borodina, T.; Markvicheva, E.; Kunizhev, S.; Möhwald, H.; Sukhorukov Gleb, B.; Kreft, O. Controlled release of DNA from self-degrading microcapsules. Macromol. Rapid Commun. 2007, 28, 1894–1899. [Google Scholar] [CrossRef]
- Li, L.; Puhl, S.; Meinel, L.; Germershaus, O. Silk fibroin layer-by-layer microcapsules for localized gene delivery. Biomaterials 2014, 35, 7929–7939. [Google Scholar] [CrossRef]
- Deng, C.X.; Sieling, F.; Pan, H.; Cui, J. Ultrasound-induced cell membrane porosity. Ultrasound Med. Biol. 2004, 30, 519–526. [Google Scholar] [CrossRef]
- Pan, H.; Zhou, Y.; Izadnegahdar, O.; Cui, J.; Deng, C.X. Study of sonoporation dynamics affected by ultrasound duty cycle. Ultrasound Med. Biol. 2005, 31, 849–856. [Google Scholar] [CrossRef]
- Zhou, Y.; Kumon, R.E.; Cui, J.; Deng, C.X. The size of sonoporation pores on the cell membrane. Ultrasound Med. Biol. 2009, 35, 1756–1760. [Google Scholar] [CrossRef]
- Zarnitsyn, V.; Rostad, C.A.; Prausnitz, M.R. Modeling transmembrane transport through cell membrane wounds created by acoustic cavitation. Biophys. J. 2008, 95, 4124–4138. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Kozlovskaya, V.; Liu, F.; Chen, J.; Williams, J.F.; Campos-Gomez, J.; Saeed, M.; Kharlampieva, E. Intracellular degradable hydrogel cubes and spheres for anti-cancer drug delivery. ACS Appl. Mater. Interfaces 2015, 7, 13633–13644. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Wang, W.; Qin, J.-J.; Nijampatnam, B.; Murugesan, S.; Kozlovskaya, V.; Zhang, R.; Velu, S.E.; Kharlampieva, E. Highly efficient delivery of potent anticancer iminoquinone derivative by multilayer hydrogel cubes. Acta Biomater. 2017, 58, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ratnayaka, S.; Alford, A.; Kozlovskaya, V.; Liu, F.; Xue, B.; Hoyt, K.; Kharlampieva, E. Theranostic multilayer capsules for ultrasound imaging and guided drug delivery. ACS Nano 2017, 11, 3135–3146. [Google Scholar] [CrossRef] [PubMed]
- Alford, A.; Rich, M.; Kozlovskaya, V.; Chen, J.; Sherwood, J.; Bolding, M.; Warram, J.; Bao, Y.; Kharlampieva, E. Ultrasound-triggered delivery of anticancer therapeutics from mri-visible multilayer microcapsules. Adv. Ther. 2018, 1, 1800051. [Google Scholar] [CrossRef]
- Hynynen, K. Ultrasound for drug and gene delivery to the brain. Adv. Drug Deliv. Rev. 2008, 60, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, A.; Brisken, A.F.; Francis, S.E.; Cumberland, D.C.; Crossman, D.C.; Newman, C.M. Microbubble-enhanced ultrasound for vascular gene delivery. Gene Ther. 2000, 7, 2023. [Google Scholar] [CrossRef]
- Melodelima, D.; Chapelon, J.Y.; Theillère, Y.; Cathignol, D. Combination of thermal and cavitation effects to generate deep lesions with an endocavitary applicator using a plane transducer: Ex vivo studies. Ultrasound Med. Biol. 2004, 30, 103–111. [Google Scholar] [CrossRef]
- Huynh, E.; Leung, B.Y.C.; Helfield, B.L.; Shakiba, M.; Gandier, J.-A.; Jin, C.S.; Master, E.R.; Wilson, B.C.; Goertz, D.E.; Zheng, G. In situ conversion of porphyrin microbubbles to nanoparticles for multimodality imaging. Nat. Nanotechnol. 2015, 10, 325. [Google Scholar] [CrossRef]
- Kozlovskaya, V.; Chen, J.; Zavgorodnya, O.; Hasan, M.B.; Kharlampieva, E. Multilayer hydrogel capsules of interpenetrated network for encapsulation of small molecules. Langmuir 2018, 34, 11832–11842. [Google Scholar] [CrossRef]
- Kozlovskaya, V.; Xue, B.; Lei, W.; Padgett, L.E.; Tse, H.M.; Kharlampieva, E. Hydrogen-bonded multilayers of tannic acid as mediators of t-cell immunity. Adv. Healthcare Mater. 2015, 4, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskaya, V.; Alexander, J.F.; Wang, Y.; Kuncewicz, T.; Liu, X.; Godin, B.; Kharlampieva, E. Internalization of red blood cell-mimicking hydrogel capsules with ph-triggered shape responses. ACS Nano 2014, 8, 5725–5737. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskaya, V.; Chen, J.; Tedjo, C.; Liang, X.; Campos-Gomez, J.; Oh, J.; Saeed, M.; Lungu, C.T.; Kharlampieva, E. Ph-responsive hydrogel cubes for release of doxorubicin in cancer cells. J. Mater. Chem. B 2014, 2, 2494–2507. [Google Scholar] [CrossRef]
- Liang, X.; Kozlovskaya, V.; Chen, Y.; Zavgorodnya, O.; Kharlampieva, E. Thermosensitive multilayer hydrogels of poly(N-vinylcaprolactam) as nanothin films and shaped capsules. Chem. Mater. 2012, 24, 3707–3719. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskaya, V.; Zavgorodnya, O.; Ankner, J.F.; Kharlampieva, E. Controlling internal organization of multilayer poly(methacrylic acid) hydrogels with polymer molecular weight. Macromolecules 2015, 48, 8585–8593. [Google Scholar] [CrossRef]
- Kozlovskaya, V.; Zavgorodnya, O.; Wang, Y.; Ankner, J.F.; Kharlampieva, E. Tailoring architecture of nanothin hydrogels: Effect of layering on pH-triggered swelling. ACS Macro Lett. 2013, 2, 226–229. [Google Scholar] [CrossRef]
- Taboury, J.; Liquier, J.; Taillandier, E. Characterization of DNA structures by infrared spectroscopy: Double helical forms of poly(dG-dC)·poly(dG-dC), poly(dd8G-dC)·poly(dd8G-dC), and poly(dG-dm5C)·poly(dG-dm5C). Can. J. Chem. 2011, 63, 1904–1909. [Google Scholar] [CrossRef]
- Bloomfield Victor, A. DNA condensation by multivalent cations. Biopolymers 1997, 44, 269–282. [Google Scholar] [CrossRef] [Green Version]
- Verdolino, V.; Cammi, R.; Munk, B.H.; Schlegel, H.B. Calculation of pKa values of nucleobases and the guanine oxidation products guanidinohydantoin and spiroiminodihydantoin using density functional theory and a polarizable continuum model. J. Phys. Chem. B 2008, 112, 16860–16873. [Google Scholar] [CrossRef]
- Jones, D.P.; Carlson, J.L.; Mody, V.C.; Cai, J.; Lynn, M.J.; Sternberg, P. Redox state of glutathione in human plasma. Free Radic. Biol. Med. 2000, 28, 625–635. [Google Scholar] [CrossRef]
- Meister, A.; Anderson, M.E. Glutathione. Ann. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef] [PubMed]
- Zelikin, A.N.; Becker, A.L.; Johnston, A.P.R.; Wark, K.L.; Turatti, F.; Caruso, F. A general approach for DNA encapsulation in degradable polymer microcapsules. ACS Nano 2007, 1, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shen, K. G-quadruplex DNA-based asymmetric catalysis of michael addition: Effects of sonication, ligands, and co-solvents. Biotechnol. Prog. 2016, 32, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Lepecq, J.B.; Paoletti, C. A fluorescent complex between ethidium bromide and nucleic acids: Physical—Chemical characterization. J. Mol. Biol. 1967, 27, 87–106. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alford, A.; Tucker, B.; Kozlovskaya, V.; Chen, J.; Gupta, N.; Caviedes, R.; Gearhart, J.; Graves, D.; Kharlampieva, E. Encapsulation and Ultrasound-Triggered Release of G-Quadruplex DNA in Multilayer Hydrogel Microcapsules. Polymers 2018, 10, 1342. https://doi.org/10.3390/polym10121342
Alford A, Tucker B, Kozlovskaya V, Chen J, Gupta N, Caviedes R, Gearhart J, Graves D, Kharlampieva E. Encapsulation and Ultrasound-Triggered Release of G-Quadruplex DNA in Multilayer Hydrogel Microcapsules. Polymers. 2018; 10(12):1342. https://doi.org/10.3390/polym10121342
Chicago/Turabian StyleAlford, Aaron, Brenna Tucker, Veronika Kozlovskaya, Jun Chen, Nirzari Gupta, Racquel Caviedes, Jenna Gearhart, David Graves, and Eugenia Kharlampieva. 2018. "Encapsulation and Ultrasound-Triggered Release of G-Quadruplex DNA in Multilayer Hydrogel Microcapsules" Polymers 10, no. 12: 1342. https://doi.org/10.3390/polym10121342