Characterization of the PB2 Cap Binding Domain Accelerates Inhibitor Design
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification
2.2. Crystallization
2.3. Data Collection
2.4. Data Processing and Refinement
3. Results and Discussion
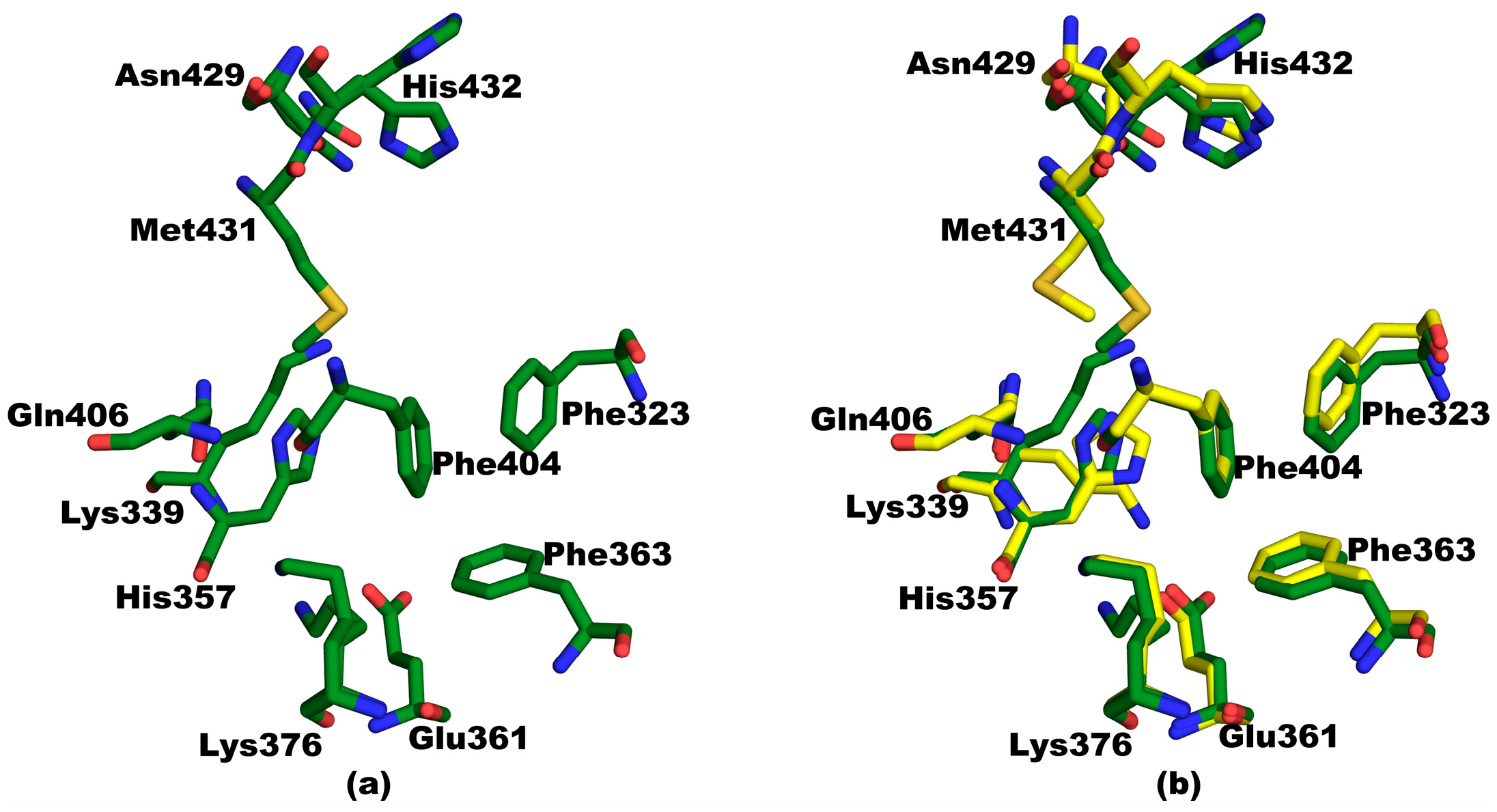
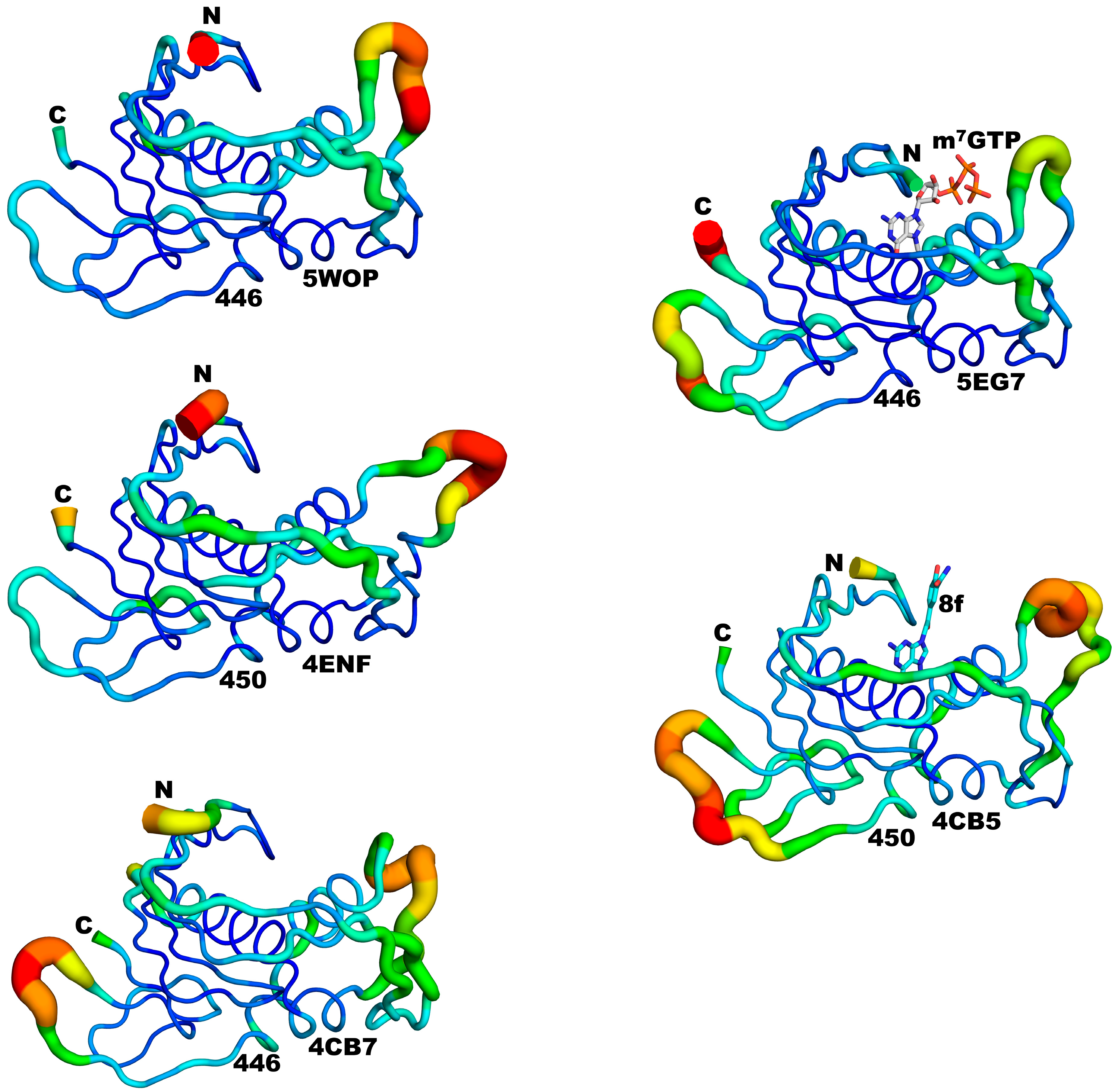
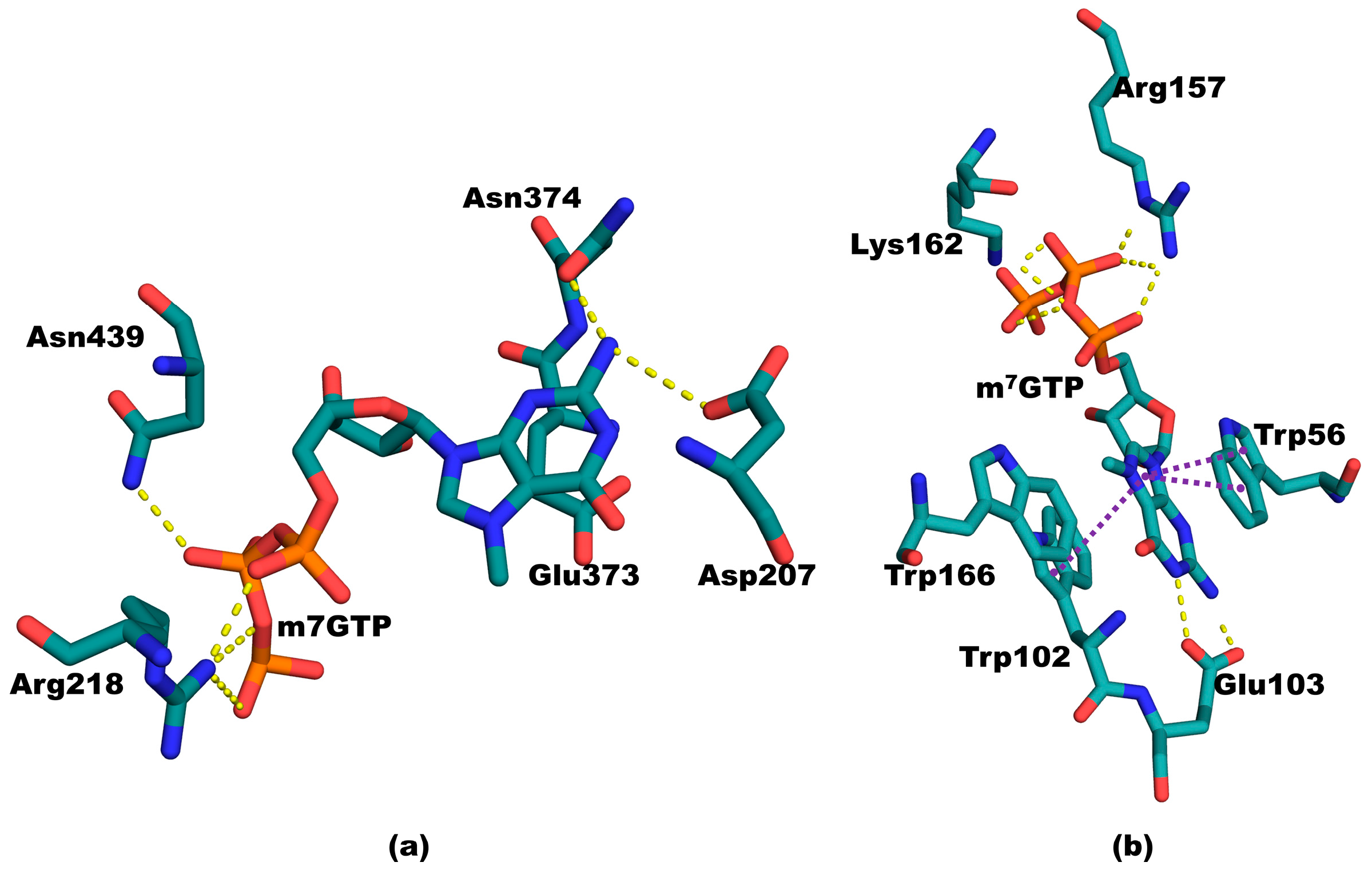
3.1. Structure of PB2cap from A/California/07/2009 (H1N1) Compared to PB2cap from A/PR/8/34 (H1N1)
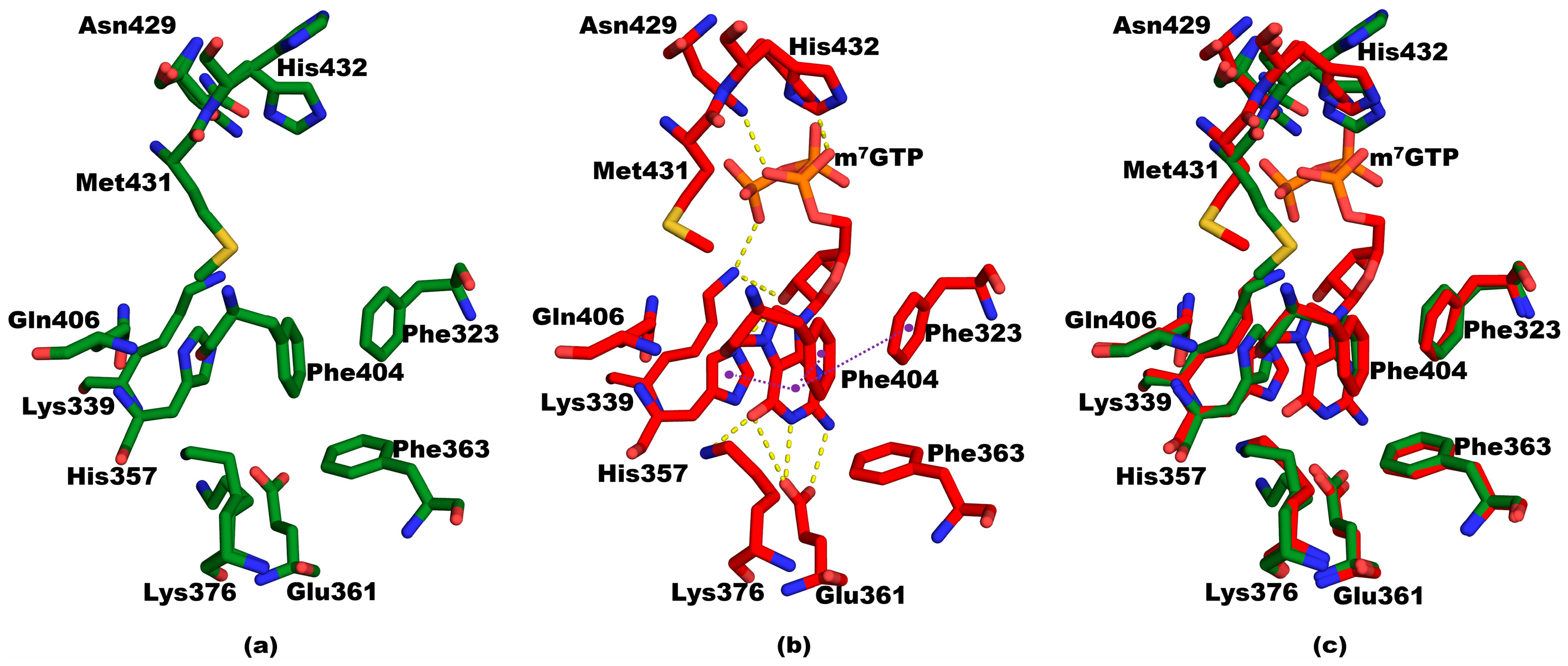
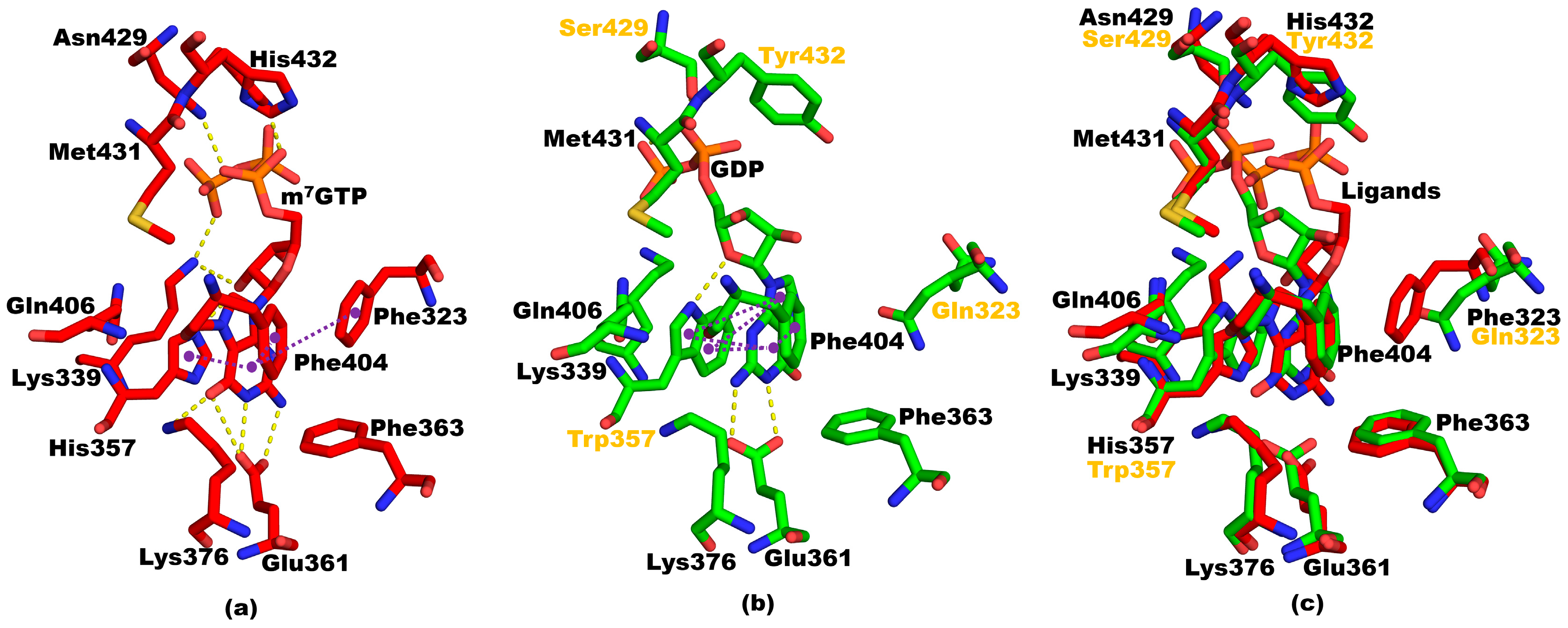
3.2. Structure of Mutant PB2cap from A/California/07/2009 (H1N1) Bound to m7GTP
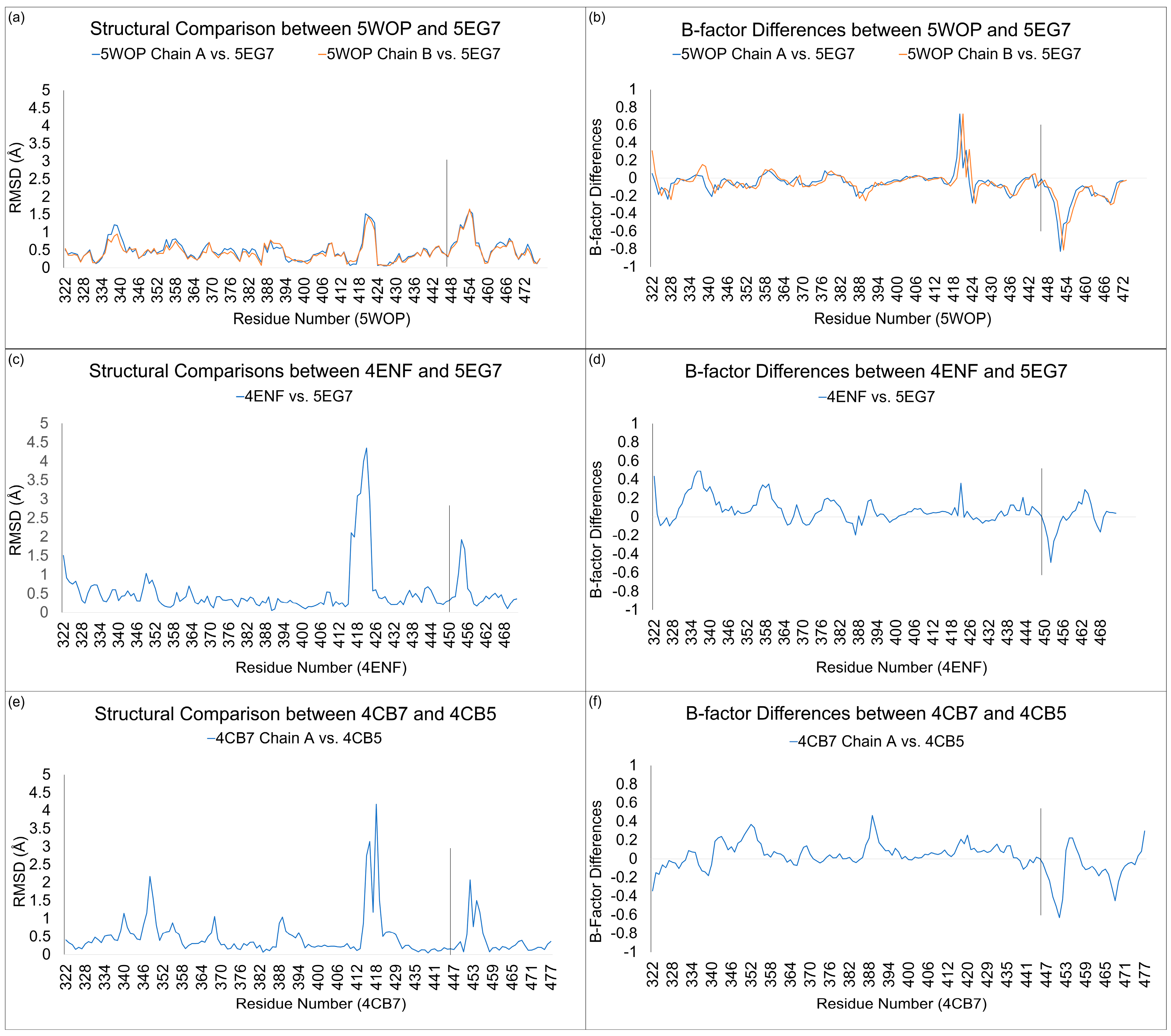
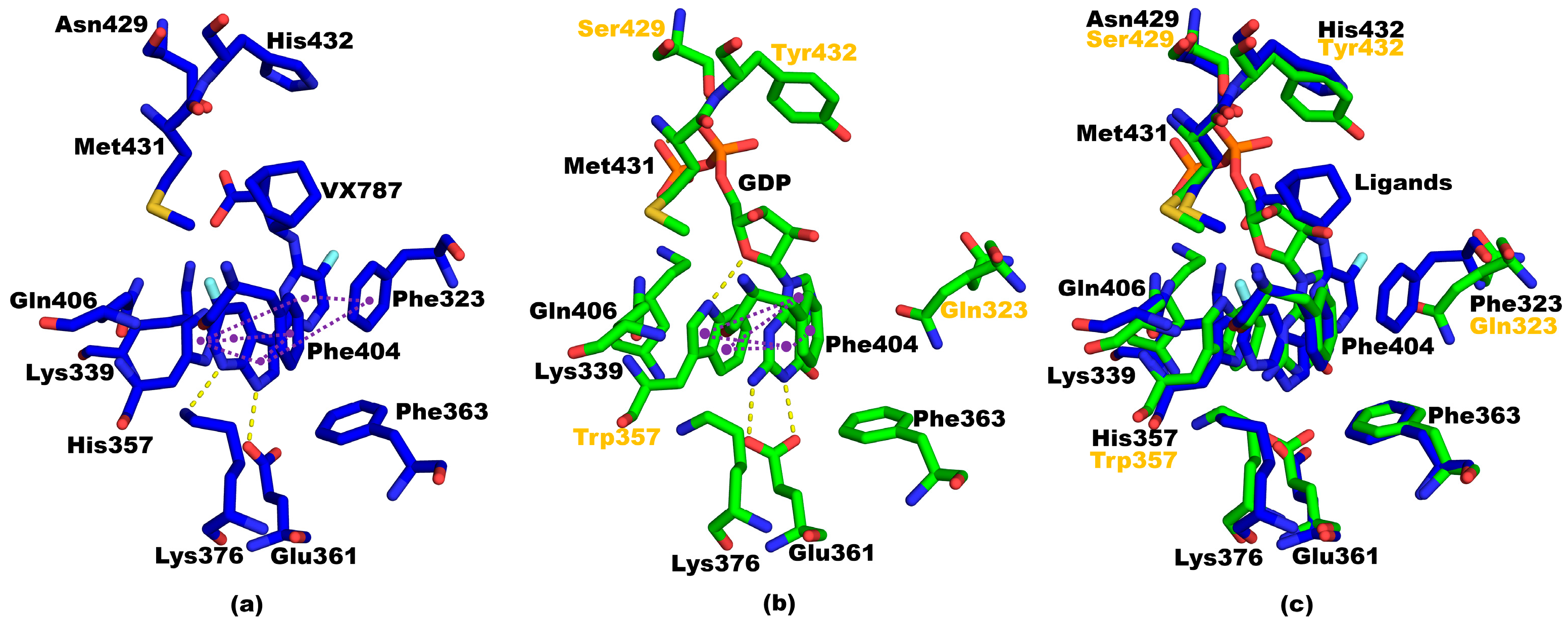
3.3. Structural Comparisons between 5EG7 and Other Structures
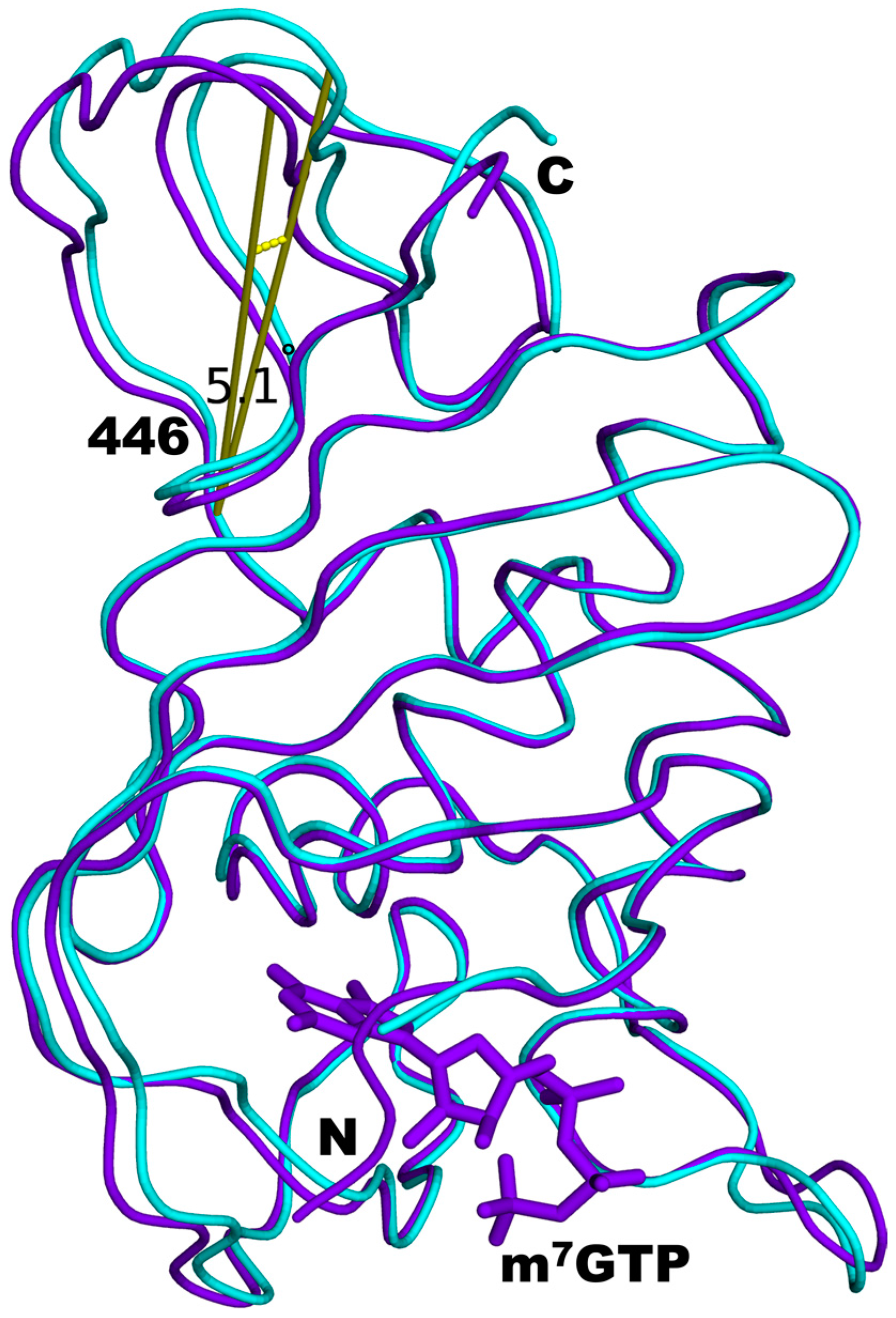
3.4. Structural Comparisons between 5EG7 and Unliganded Mutant PB2cap
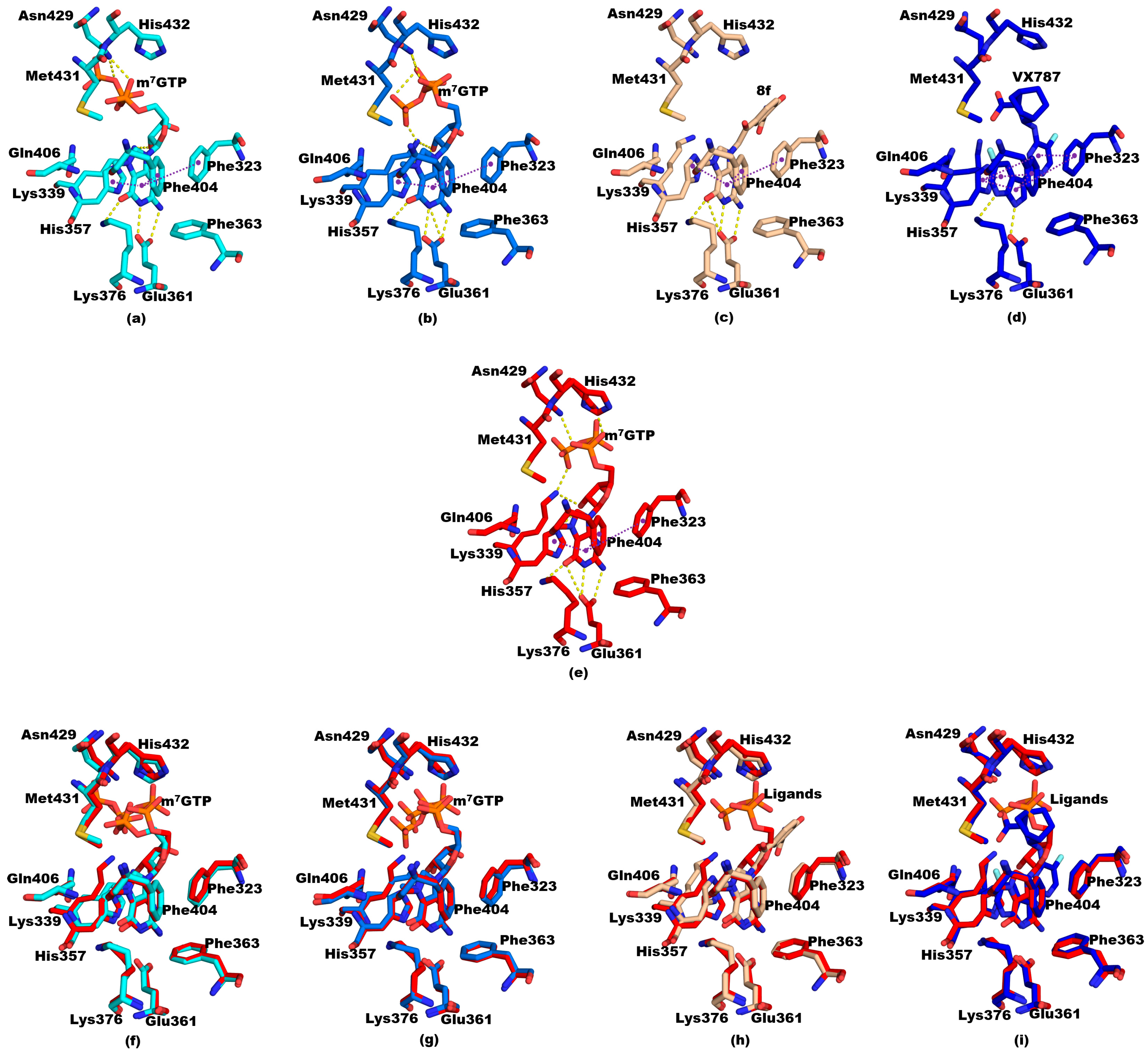
3.5. Structural Comparisons between Other PB2cap Structures
3.6. Structural Ccomparisons between 5EG7 and PB2cap from B/Jiangxi/BV/2006 PB2cap Bound to GDP
3.7. Structural Ccomparisons between 4Q46 and 4P1U 6
3.8. The Difference between PB2cap and Human Cap-Binding Proteins: Cap-Specific mRNA (Nucleoside-2′-O-)-methyltransferase 1 and Eukaryotic Translation Initiation Factor 4E
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 1999, 3, 707–716. [Google Scholar] [CrossRef]
- Mazza, C.; Segref, A.; Mattaj, I.W.; Cusack, S. Large-scale induced fit recognition of an m(7)gpppg cap analogue by the human nuclear cap-binding complex. EMBO J. 2002, 21, 5548–5557. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Rao, P.; Krug, R.M. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 2001, 20, 2078–2086. [Google Scholar] [CrossRef] [PubMed]
- Guilligay, D.; Tarendeau, F.; Resa-Infante, P.; Coloma, R.; Crepin, T.; Sehr, P.; Lewis, J.; Ruigrok, R.W.; Ortin, J.; Hart, D.J.; et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008, 15, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, K.; Obayashi, E.; Kawaguchi, A.; Suzuki, Y.; Tame, J.R.; Nagata, K.; Park, S.Y. Structural insight into the essential pb1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 2009, 28, 1803–1811. [Google Scholar] [CrossRef] [PubMed]
- Plotch, S.J.; Bouloy, M.; Ulmanen, I.; Krug, R.M. A unique cap(m7gpppxm)-dependent influenza virion endonuclease cleaves capped rnas to generate the primers that initiate viral RNA transcription. Cell 1981, 23, 847–858. [Google Scholar] [CrossRef]
- Jung, T.E.; Brownlee, G.G. A new promoter-binding site in the PB1 subunit of the influenza a virus polymerase. J. Gen. Virol. 2006, 87, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Bartlam, M.; Lou, Z.; Chen, S.; Zhou, J.; He, X.; Lv, Z.; Ge, R.; Li, X.; Deng, T.; et al. Crystal structure of an avian influenza polymerase PA(n) reveals an endonuclease active site. Nature 2009, 458, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.; Bouvier, D.; Crepin, T.; McCarthy, A.A.; Hart, D.J.; Baudin, F.; Cusack, S.; Ruigrok, R.W. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Severin, C.; Rocha de Moura, T.; Liu, Y.; Li, K.; Zheng, X.; Luo, M. The cap-binding site of influenza virus protein PB2 as a drug target. Acta Crystallogr. D Struct. Biol. 2016, 72, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qin, K.; Meng, G.; Zhang, J.; Zhou, J.; Zhao, G.; Luo, M.; Zheng, X. Structural and functional characterization of k339t substitution identified in the PB2 subunit cap-binding pocket of influenza a virus. J. Biol. Chem. 2013, 288, 11013–11023. [Google Scholar] [CrossRef] [PubMed]
- Pautus, S.; Sehr, P.; Lewis, J.; Fortune, A.; Wolkerstorfer, A.; Szolar, O.; Guilligay, D.; Lunardi, T.; Decout, J.L.; Cusack, S. New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J. Med. Chem. 2013, 56, 8915–8930. [Google Scholar] [CrossRef] [PubMed]
- Constantinides, A.; Gumpper, R.H.; Severin, C.; Luo, M. High-resolution structure of influenza a PB2cap binding domain illuminates changes induced by ligand binding. Acta Crystallogr. F 2018. Accepted. [Google Scholar]
- Clark, M.P.; Ledeboer, M.W.; Davies, I.; Byrn, R.A.; Jones, S.M.; Perola, E.; Tsai, A.; Jacobs, M.; Nti-Addae, K.; Bandarage, U.K.; et al. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (vx-787) of influenza PB2. J. Med. Chem. 2014, 57, 6668–6678. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Y.; Fan, J.; He, R.; Luo, M.; Zheng, X. The crystal structure of the PB2 cap-binding domain of influenza b virus reveals a novel cap recognition mechanism. J. Biol. Chem. 2015, 290, 9141–9149. [Google Scholar] [CrossRef] [PubMed]
- Smietanski, M.; Werner, M.; Purta, E.; Kaminska, K.H.; Stepinski, J.; Darzynkiewicz, E.; Nowotny, M.; Bujnicki, J.M. Structural analysis of human 2′-o-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 2014, 5, 3004. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, E.; Jenni, S.; Kabha, E.; Takrouri, K.J.; Yi, T.; Salvi, N.; Luna, R.E.; Gavathiotis, E.; Mahalingam, P.; Arthanari, H.; et al. Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EIG-1 reveals an allosteric mechanism for dissociating eIF4G. Proc. Natl. Acad. Sci. USA 2014, 111, E3187–E3195. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- The Pymol Molecular Graphics System, version 1.3; Schrödinger, LLC: New York, NY, USA, 2010.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. Biovia Discovery Studio 2017 r2: A Comprehensive Predictive Science Application for the Life Sciences. Available online: http://accelrys.com/products/collaborative-science/biovia-discovery-studio (accessed on 29 December 2017).
- The Pymol Molecular Graphics System, version 1.8; Schrödinger, LLC: New York, NY, USA, 2015.
- Reich, S.; Guilligay, D.; Pflug, A.; Malet, H.; Berger, I.; Crepin, T.; Hart, D.; Lunardi, T.; Nanao, M.; Ruigrok, R.W.; et al. Structural insight into cap-snatching and rna synthesis by influenza polymerase. Nature 2014, 516, 361–366. [Google Scholar] [CrossRef] [PubMed]

| Protein | CA09-PB2cap | m7GTP-Mutant CA09-PB2cap | Mutant CA09-PB2cap |
|---|---|---|---|
| PDB code | 5EG8 | 5EG7 | 5WOP |
| Data collection | |||
| Space-group | P3121 | P43212 | P1211 |
| Unit-cell parameters | |||
| a (Å) | 54.61 | 45.26 | 36.86 |
| b (Å) | 54.61 | 45.26 | 72.43 |
| c (Å) | 196.49 | 156.4 | 61.32 |
| α (°) | 90 | 90 | 90 |
| β (°) | 90 | 90 | 103.18 |
| γ (°) | 120 | 90 | 90 |
| Molecules in asymmetric unit | 2 | 1 | 2 |
| Wavelength (Å) | 0.9795 | 1 | 1 |
| Resolution (Å) | 15–1.54 (1.59–1.54) | 25–1.40 (1.42–1.40) | 36.21–1.52 (1.574–1.52) |
| Rmerge † (%) | 7.0 (21.4) | 7.1 (61.1) | 4.6 (47.3) |
| 〈I/σ(I)〉 | 57.9 (14.2) | 33.2 (1.8) | 11.05 (2.03) |
| CC1/2 | 0.997 (0.992) | 0.912 (0.581) | 0.998 (0.806) |
| Completeness (%) | 97.2 (96.9) | 99.8 (97.0) | 99.17 (99.94) |
| Multiplicity | 15.1 (16.7) | 10.4 (4.0) | 2.0 (2.0) |
| Refinement | |||
| Resolution (Å) | 14.63–1.54 | 25–1.40 | 36.21–1.52 |
| No. of reflections | 47,547 | 31,406 | 93,971 |
| Rwork/Rfree ‡ (%) | 12.2/17.7 | 12.4/16.9 | 0.1498/0.2044 |
| Average B factor (Å2) | 24.9 | 16.1 | 22.01 |
| R.m.s. deviations | |||
| Bond lengths (°) | 0.019 | 0.02 | 0.026 |
| Bond angles (°) | 1.93 | 2.35 | 2.22 |
| Ramachandran plot (%) | |||
| Favored region | 97.5 | 98.1 | 98.09 |
| Allowed region | 2.2 | 1.9 | 1.91 |
| Outlier region § | 0.3 | — | 0 |
| Structures in Comparison | N-Subdomain RMSD (Å) | C-Subdomain RMSD (Å) | C†-Subdomain RMSD (Å) | C†-Subdomain Angular Distance (°) |
|---|---|---|---|---|
| 5WOP Chain A vs. 5EG7 | 0.39 | 0.40 | 1.10 | 5.1 |
| 5WOP Chain B vs. 5EG7 | 0.36 | 0.40 | 1.06 | 4.7 |
| 4ENF vs. 5EG7 | 0.55 | 0.49 | 0.63 | 4.8 |
| 4CB7 Chain A vs. 4CB5 | 0.50 | 0.33 | 0.53 | 3.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Constantinides, A.E.; Severin, C.C.; Gumpper, R.H.; Zheng, X.; Luo, M. Characterization of the PB2 Cap Binding Domain Accelerates Inhibitor Design. Crystals 2018, 8, 62. https://doi.org/10.3390/cryst8020062
Constantinides AE, Severin CC, Gumpper RH, Zheng X, Luo M. Characterization of the PB2 Cap Binding Domain Accelerates Inhibitor Design. Crystals. 2018; 8(2):62. https://doi.org/10.3390/cryst8020062
Chicago/Turabian StyleConstantinides, Amanda E., Chelsea C. Severin, Ryan H. Gumpper, Xiaofeng Zheng, and Ming Luo. 2018. "Characterization of the PB2 Cap Binding Domain Accelerates Inhibitor Design" Crystals 8, no. 2: 62. https://doi.org/10.3390/cryst8020062





