Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phylogenetic Analysis
2.2. Functional Study
2.3. Characteristics of Enzyme Activity
2.4. Thermal Stability
2.5. Structure Analysis
3. Material and Methods
3.1. Site-Directed Mutagenesis, Cloning and Expression of NDM Variants
3.2. Antimicrobial Susceptibility Tests
3.3. Production and Purification of NDM-1, NDM-5, and NDM-24
3.4. Determination of Kinetic Parameters
3.5. Circular Dichroism and Structure Analysis
3.6. Thermal Stability Testing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Palzkill, T. Metallo-beta-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Garau, G.; García-Sáez, I.; Bebrone, C.; Anne, C.; Mercuri, P.; Galleni, M.; Frère, J.M.; Dideberg, O. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–2349. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo-β-Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Poirel, L.; Carattoli, A.; Nordmann, P. Characterization of an IncFII Plasmid Encoding NDM-1 from Escherichia coli ST131. PLoS ONE 2012, 7, 34752. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Villa, L.; Poirel, L.; Nordmann, P.; Carattoli, A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 2013, 68, 34–39. [Google Scholar] [CrossRef] [PubMed]
- King, D.; Strynadka, N. Crystal structure of New Delhi metallo-betalactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.J.; Bahr, G.; Nakashige, T.G.; Nolan, E.M.; Bonomo, R.A.; Vila, A.J. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat. Chem. Biol. 2016, 12, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.J.; Bahr, G.; Vila, A.J. Lipidated beta-lactamases: From bench to bedside. Future Microbiol. 2016, 11, 1495–1498. [Google Scholar] [CrossRef]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, genetics and worldwide spread of New Delhi metallo-beta-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Cheng, Z.; Thomas, P.W.; Ju, L.; Bergstrom, A.; Mason, K.; Clayton, D.; Miller, C.; Bethel, C.R.; Vanpelt, J.; Tierney, D.L.; et al. Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: Effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J. Biol. Chem. 2018, 293, 12606–12618. [Google Scholar] [CrossRef]
- Zhang, H.O.; Hau, Q. Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J. 2011, 25, 2574–2582. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cunningham, M.A.; Mire, J.; Tesar, C.; Sacchettini, J.; Joachimiak, A. NDM-1, the ultimate promiscuous enzyme: Substrate recognition and catalytic mechanism. FASEB J. 2013, 27, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, Y.; McNally, A.; Zong, Z. bla NDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J. Antimicrob. Chemother. 2018, 73, 2336–2339. [Google Scholar] [CrossRef] [PubMed]
- Bahr, G.; Vitor-Horen, L.; Bethel, C.R.; Bonomo, R.A.; González, L.J.; Vila, A.J. Clinical evolution of New Delhi Metallo-β-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob. Agents Chemother. 2018, 62, 1817–1849. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, J.; Wang, X.; Liu, D.; Ke, Y.; Wang, Y.; Shen, J. Novel Variant of New Delhi Metallo-β-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 2018, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Walsh, T.R.; Liu, D.; Shen, Z.; Zhang, R.; Yin, W.; Yao, H.; Li, J.; Shen, J. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a ST48 Escherichia coli strain. Antimicrob. Agents Chemother. 2017, 61, 2216–2233. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Boulanger, A.E.; Poirel, L. NDM-4 Metallo-β-Lactamase with Increased Carbapenemase Activity from Escherichia coli. Antimicrob. Agents Chemother. 2012, 56, 2184–2186. [Google Scholar] [CrossRef]
- Zou, D.; Huang, Y.; Zhao, X.; Liu, W.; Dong, D.; Li, H.; Wang, X.; Huang, S.; Wei, X.; Yan, X.; et al. A Novel New Delhi Metallo-β-Lactamase Variant, NDM-14, Isolated in a Chinese Hospital Possesses Increased Enzymatic Activity against Carbapenems. Antimicrob. Agents Chemother. 2015, 59, 2450–2453. [Google Scholar] [CrossRef]
- Tada, T.; Miyoshi-Akiyama, T.; Dahal, R.K.; Sah, M.K.; Ohara, H.; Kirikae, T.; Pokhrel, B.M. NDM-8 Metallo-β-Lactamase in a Multidrug-Resistant Escherichia coli Strain Isolated in Nepal. Antimicrob. Agents Chemother. 2013, 57, 2394–2396. [Google Scholar] [CrossRef]
- Tada, T.; Shrestha, B.; Miyoshi-Akiyama, T.; Shimada, K.; Ohara, H.; Kirikae, T.; Pokhrel, B.M. NDM-12, a Novel New Delhi Metallo-β-Lactamase Variant from a Carbapenem-Resistant Escherichia coli Clinical Isolate in Nepal. Antimicrob. Agents Chemother. 2014, 58, 6302–6305. [Google Scholar] [CrossRef]
- Ines, S.; Emma, K.; Rudolf, R.; Rumyana, M.; Anne Marie, Q.; Adolf, B. VIM-15 and VIM-16, two new VIM-2-like metallo-beta-lactamases in Pseudomonas aeruginosa isolates from Bulgaria and Germany. Antimicrob. Agents Chemother. 2008, 52, 2977. [Google Scholar]
- Patricia, M.; Tomatis, P.E.; Mussi, M.A.; Fernando, P.; Viale, A.M.; Limansky, A.S.; Vila, A.J. Biochemical characterization of metallo-beta-lactamase VIM-11 from a Pseudomonas aeruginosa clinical strain. Antimicrob. Agents Chemother. 2008, 52, 2250. [Google Scholar]
- Jose-Manuel, R.M.; Patrice, N.; Nicolas, F.; Laurent, P. VIM-19, a metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2010, 54, 471–476. [Google Scholar]
- Pierre, B.; Carine, B.; Te-Din, H.; Warda, B.; Yves, D.; Ariane, D.; Kurt, H.; Youri, G. Detection and characterization of VIM-31, a new variant of VIM-2 with Tyr224His and His252Arg mutations, in a clinical isolate of Enterobacter cloacae. Antimicrob. Agents Chemother. 2012, 56, 3283. [Google Scholar]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Issa, B.; Kar, D.; Biswal, S.; Ghosh, A.S. E152A substitution drastically affects NDM-5 activity. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Azam, M.W.; Khan, A.U. Non-active site mutation (Q123A) in New Delhi metallo-β-lactamase (NDM-1) enhanced its enzyme activity. Int. J. Biol. Macromol. 2018, 112, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Carfi, A.; Pares, S.; Duée, E.; Galleni, M.; Duez, C.; Frère, J.M.; Dideberg, O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921. [Google Scholar] [CrossRef] [PubMed]
- Piccirilli, A.; Brisdelli, F.; Aschi, M.; Celenza, G.; Amicosante, G.; Perilli, M. Kinetic Profile and Molecular Dynamic Studies Show that Y229W Substitution in an NDM-1/L209F Variant Restores the Hydrolytic Activity of the Enzyme toward Penicillins, Cephalosporins, and Carbapenems. Antimicrob. Agents Chemother. 2019, 63, e02270-18. [Google Scholar] [CrossRef] [Green Version]
- Meini, M.-R.; Tomatis, P.E.; Weinreich, D.M.; Vila, A.J. Quantitative Description of a Protein Fitness Landscape Based on Molecular Features. Mol. Boil. Evol. 2015, 32, 1774–1787. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard, 11th ed.; CLSI document M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI document M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Crowder, M.W.; Walsh, T.R.; Banovic, L.; Pettit, M.; Spencer, J. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 1998, 42, 921. [Google Scholar] [CrossRef] [PubMed]
- De Meester, F.; Joris, B.; Reckinger, G. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem. Pharmacol. 1987, 36, 2393–2403. [Google Scholar] [CrossRef]
- Segel, I.H. Biochemical Calculations, 2nd ed.; John Wiley & Sons: New York, NY, USA, 1976; pp. 236–241. [Google Scholar]
- Liu, Z.; Zhang, R.; Li, W.; Yang, L.; Liu, D.; Wang, S.; Shen, J.; Wang, Y. Amino acid changes at the VIM-48 C-terminus result in increased carbapenem resistance, enzyme activity and protein stability. J. Antimicrob. Chemother. 2018, 74, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Raussens, V.; Ruysschaert, J.-M.; Goormaghtigh, E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: A new scaling method. Anal. Biochem. 2003, 319, 114–121. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999, 8, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Analysis of Protein Circular Dichroism Spectra Based on the Tertiary Structure Classification. Anal. Biochem. 2001, 299, 271–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]


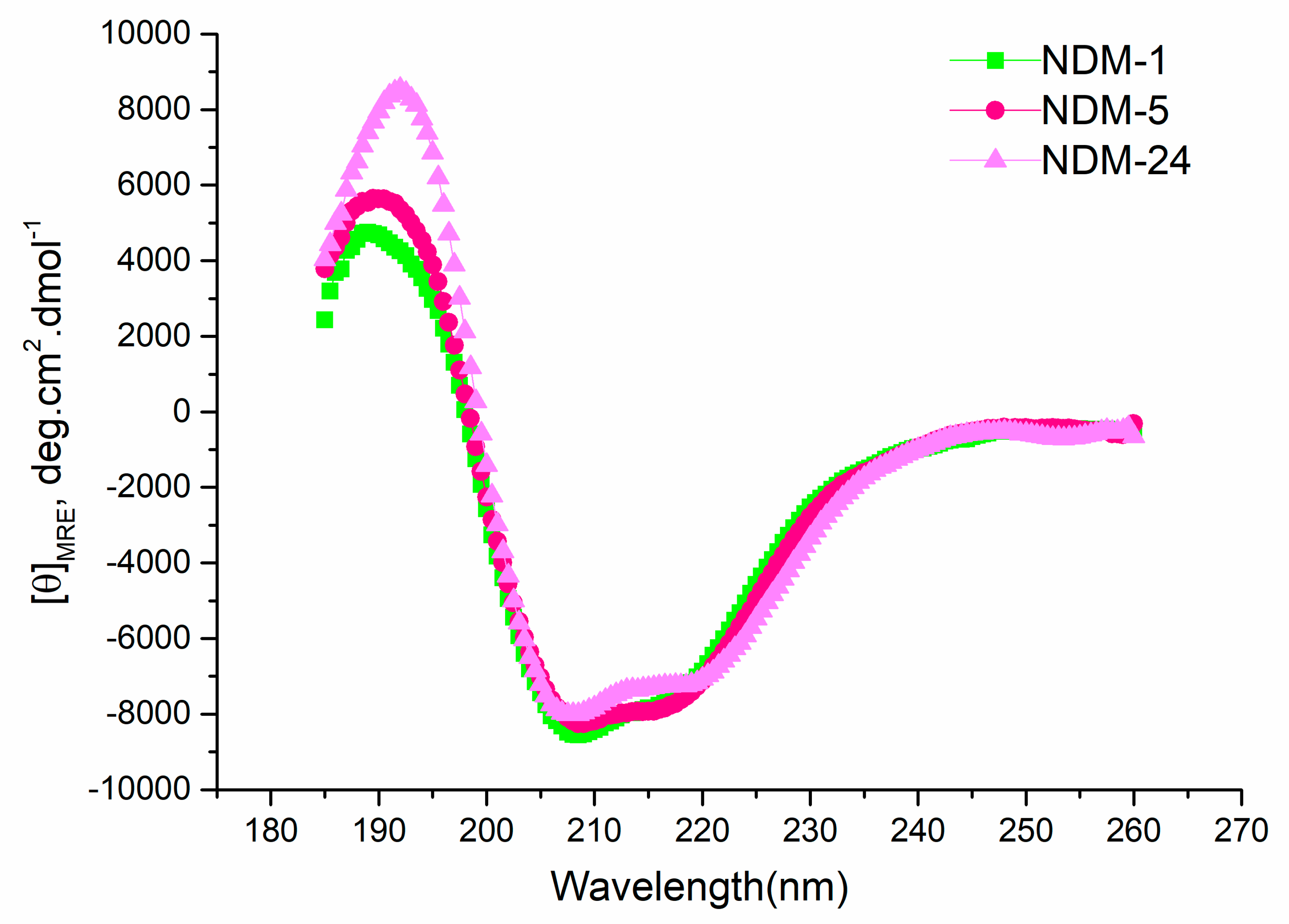
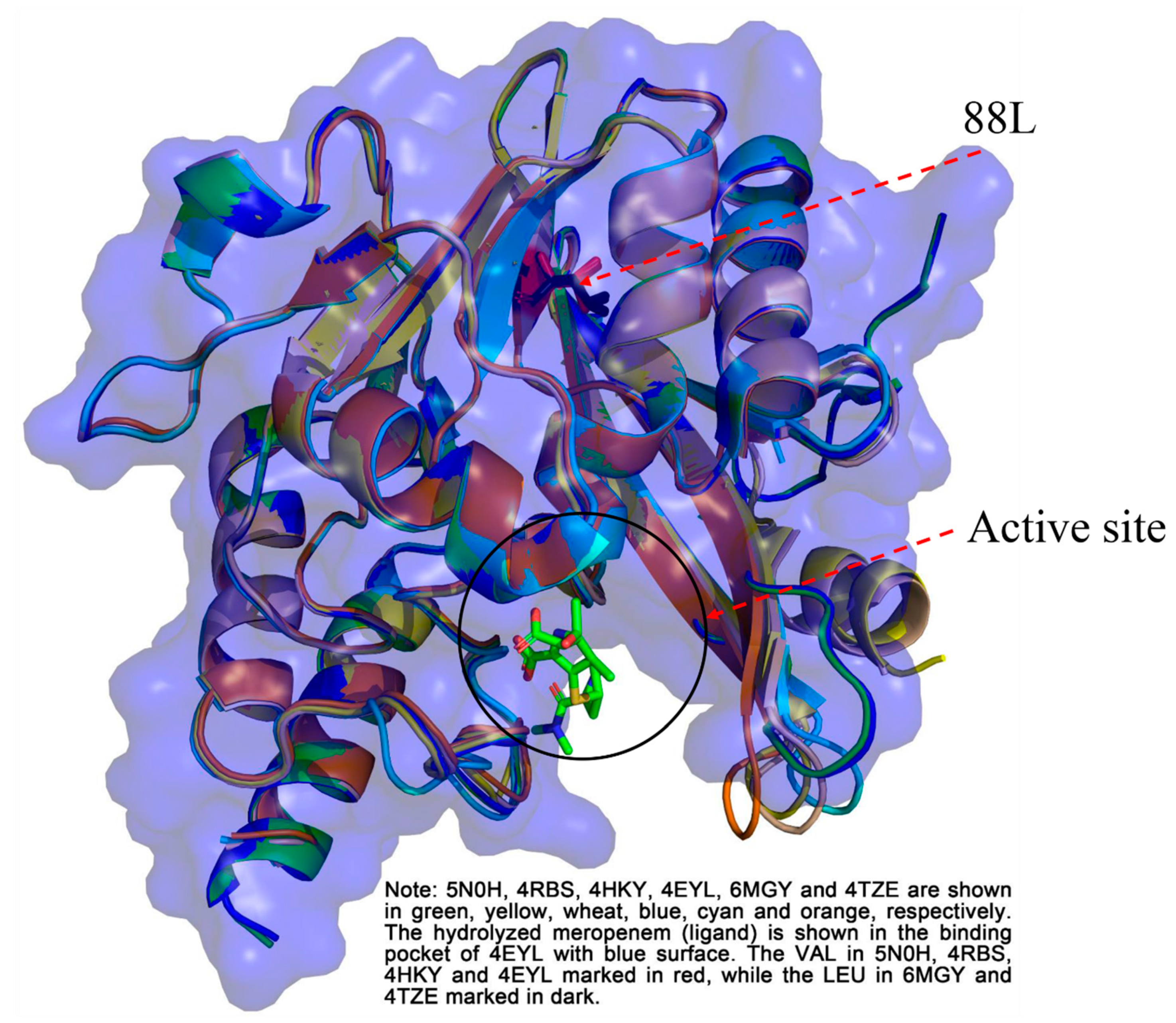
| Antibiotic | MIC (μg/mL) | |||
|---|---|---|---|---|
| E. coli DH5α/pHSG398 | E. coli DH5α/pHSG398-NDM-24 | E. coli DH5α/pHSG398-NDM-1 | E. coli DH5α/pHSG398-NDM-5 | |
| Ampicillin | 2 | >256 | >256 | >256 |
| Penicillin G | 16 | >256 | >256 | >256 |
| Aztreonam | 0.031 | 0.031 | 0.031 | 0.031 |
| Cefepime | 0.031 | 2 | 1 | 2 |
| Cefotaxime | 0.062 | 32 | 64 | 64 |
| Cefoxitin | 2 | 128 | 128 | 128 |
| Ceftazidime | 0.125 | 256 | 256 | 256 |
| Cefazolin | 2 | 128 | 128 | 256 |
| Ertapenem | 0.015 | 1 | 0.25 | 2 |
| Imipenem | 0.062 | 2 | 2 | 2 |
| Meropenem | 0.031 | 1 | 1 | 2 |
| Kinetic Parameters | Enzyme | β-Lactams b | ||||||
|---|---|---|---|---|---|---|---|---|
| AMP | PEN | TAG | FEP | MEM | IPM | ETP | ||
| Km(μM) | NDM-24 | 638.79 ± 23.86 | 331.30 ± 29.43 | 173.85 ± 9.73 | 318.93 ± 10.86 | 266.24 ± 27.03 | 338.20 ± 24.23 | 125.23 ± 19.08 |
| NDM-1 | 1249.98 ± 210.94 | 224.57 ± 13.57 | 213.90 ± 11.01 | 173.55 ± 19.46 | 284.24 ± 7.87 | 234.83 ± 7.44 | 105.54 ± 3.09 | |
| NDM-5 | 825.00 ± 0.29 | 315.21 ± 46.68 | 76.45 ± 4.76 | 179.64 ± 12.19 | 275.16 ± 36.87 | 292.97 ± 13.76 | 82.18 ± 3.86 | |
| kcat (s−1) | NDM-24 | 259.94 ± 23.52 | 179.10 ± 8.17 | 43.13 ± 1.06 | 22.98 ± 0.34 | 151.75 ± 6.69 | 173.16 ± 8.83 | 110.31 ± 7.62 |
| NDM-1 | 254.34 ± 28.96 | 79.28 ± 1.96 | 26.73 ± 0.71 | 8.42 ± 0.63 | 75.18 ± 3.44 | 79.81 ± 5.15 | 62.89 ± 1.15 | |
| NDM-5 | 346.13 ± 31.30 | 214.13 ± 12.11 | 26.96 ± 0.75 | 13.05 ± 0.24 | 142.48 ± 17.91 | 149.63 ± 2.02 | 83.18 ± 1.67 | |
| kcat/Km (μM−1 s−1) | NDM-24 | 0.41 | 0.54 | 0.25 | 0.072 | 0.57 | 0.51 | 0.88 |
| NDM-1 | 0.20 | 0.35 | 0.13 | 0.046 | 0.26 | 0.34 | 0.60 | |
| NDM-5 | 0.40 | 0.68 | 0.35 | 0.073 | 0.52 | 0.51 | 1.01 | |
| kcat/Km (μM−1 s−1) ratio for: | NDM-24/NDM-1 | 2.00 | 1.53 | 1.98 | 1.49 | 2.16 | 1.51 | 1.46 |
| NDM-5/NDM-24 | 1.03 | 1.26 | 1.42 | 1.01 | 0.91 | 1.00 | 1.15 | |
| NDM-5/NDM-1 | 2.07 | 1.92 | 2.82 | 1.50 | 1.96 | 1.50 | 1.68 | |
| Program Algorithms a | Structural Elements b | SMP50(9) c | SP37(3) c | SP29(1) c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NDM-1 | NDM-5 | NDM-24 | NDM-1 | NDM-5 | NDM-24 | NDM-1 | NDM-5 | NDM-24 | ||
| SELCON3 | H(r) | 0.070 | 0.078 | 0.092 | 0.062 | 0.074 | 0.092 | 0.059 | 0.079 | 0.087 |
| H(d) | 0.085 | 0.088 | 0.089 | 0.081 | 0.088 | 0.089 | 0.078 | 0.087 | 0.086 | |
| S(r) | 0.215 | 0.199 | 0.195 | 0.228 | 0.214 | 0.195 | 0.231 | 0.191 | 0.196 | |
| S(d) | 0.115 | 0.109 | 0.108 | 0.117 | 0.113 | 0.108 | 0.118 | 0.107 | 0.108 | |
| Trn | 0.214 | 0.211 | 0.194 | 0.218 | 0.214 | 0.194 | 0.226 | 0.214 | 0.215 | |
| Unrd | 0.284 | 0.287 | 0.261 | 0.282 | 0.279 | 0.261 | 0.287 | 0.292 | 0.285 | |
| H(r)+H(d) | 0.155 | 0.166 | 0.181 | 0.143 | 0.162 | 0.181 | 0.137 | 0.166 | 0.173 | |
| S(r)+S(d) | 0.33 | 0.308 | 0.303 | 0.345 | 0.327 | 0.303 | 0.349 | 0.298 | 0.304 | |
| CONTINLL | H(r) | 0.054 | 0.075 | 0.091 | 0.046 | 0.079 | 0.097 | 0.071 | 0.078 | 0.093 |
| H(d) | 0.079 | 0.092 | 0.101 | 0.089 | 0.095 | 0.103 | 0.092 | 0.096 | 0.100 | |
| S(r) | 0.217 | 0.208 | 0.187 | 0.202 | 0.205 | 0.182 | 0.197 | 0.197 | 0.183 | |
| S(d) | 0.114 | 0.113 | 0.108 | 0.112 | 0.111 | 0.107 | 0.113 | 0.111 | 0.107 | |
| Trn | 0.233 | 0.220 | 0.220 | 0.248 | 0.216 | 0.216 | 0.231 | 0.222 | 0.225 | |
| Unrd | 0.303 | 0.292 | 0.293 | 0.304 | 0.293 | 0.294 | 0.297 | 0.297 | 0.292 | |
| H(r)+H(d) | 0.133 | 0.167 | 0.192 | 0.135 | 0.174 | 0.200 | 0.163 | 0.174 | 0.193 | |
| S(r)+S(d) | 0.331 | 0.321 | 0.295 | 0.314 | 0.316 | 0.289 | 0.310 | 0.308 | 0.290 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Piccirilli, A.; Liu, D.; Li, W.; Wang, Y.; Shen, J. Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase. Catalysts 2019, 9, 744. https://doi.org/10.3390/catal9090744
Liu Z, Piccirilli A, Liu D, Li W, Wang Y, Shen J. Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase. Catalysts. 2019; 9(9):744. https://doi.org/10.3390/catal9090744
Chicago/Turabian StyleLiu, Zhihai, Alessandra Piccirilli, Dejun Liu, Wan Li, Yang Wang, and Jianzhong Shen. 2019. "Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase" Catalysts 9, no. 9: 744. https://doi.org/10.3390/catal9090744





