Identification of Key Amino Acid Residues Determining Product Specificity of 2,3-Oxidosqualene Cyclase in Siraitia grosvenorii
Abstract
:1. Introduction
2. Results
2.1. Catalytic Mechanism Prediction of Cucurbitadienol Biosynthesis in S. grosvenorii
2.2. Bioinformatic Analysis of SgCS
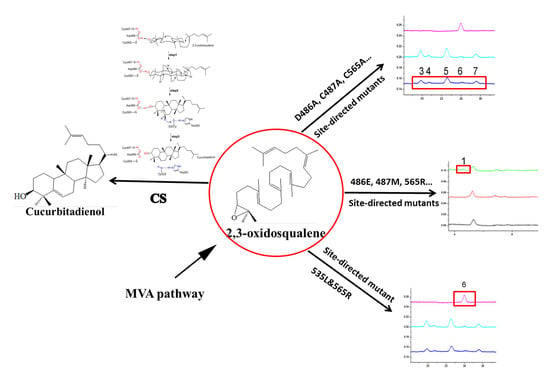
2.3. DCTAE Motif for the Initiation of the Polycyclization Cascade
2.4. Functions of the Tyr535 and His260 Residue
2.5. Double Mutants Determining Product Specificity
2.6. Reaction Mechanism of SgCS
3. Discussion
4. Materials and Methods
4.1. Chemicals and General Methods
4.2. Molecular Modeling
4.3. Molecular Docking
4.4. Mutagenesis Experiments of SgCS
4.5. Yeast Transformation and Cell Cultivation
4.6. UPLC-Q-TOF Analysis of Yeast Extracts
4.7. Compound Purification from Yeast
4.8. Compound Structure Elucidation by NMR Data Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Protein Characteristic | SgCS |
|---|---|
| Number of the deduced amino acids | 759 |
| Molecular weight (kDa) | 86.4 |
| Theoretical pI | 6.64 |
| Instability index | 38.07 |
| Grand average of hydropathicity (GRAVY) | −0.313 |
| Mutants | Primers (5′→3′) |
|---|---|
| USER cassette oligo for pCEV-G4-Km, SpeI/PacI overhang. | F: CTAGTGCTGAGGCATGTTTAAACGACCCTCAGCTTAAT R: TAAGCTGAGGGTCGTTTAAACATGCCTCAGCA |
| SgCS-Wild | F: GGCATGTTUATGTGGAGATTGAAAGTTGGTGC R: GGGTCGTTUTCACTCTGTTAAAACTCTGTGGCAGTAC |
| pCEV-Seq | F: CGATGACCTCCCATTGATA R: CGTTCTTAATACTAACATAACT |
| D486E | F: AGATCACGGTTGGTTAATTTCTGAAGTACTGCTGAAGGTTTAAAGGC R: GCCTTTAAACCTTCAGCAGTACTTCAGAAATTAACCAACCGTGATCT |
| D486N | F: AGATCACGGTTGGTTAATTTCTAACGTACTGCTGAAGGTTTAAAGGC R: GCCTTTAAACCTTCAGCAGTACGTTAGAAATTAACCAACCGTGATCT |
| D486A | F: AGATCACGGTTGGTTAATTTCTGCTGTACTGCTGAAGGTTTAAAGGC R: GCCTTTAAACCTTCAGCAGTACAGCAGAAATTAACCAACCGTGATCT |
| C487M | F: GACTACCCTTACGTTGAGATGACATCTGCAACAATGGAGG R: CCTCCATTGTTGCAGATGTCATCTCAACGTAAGGGTAGTC |
| C487R | F: GACTACCCTTACGTTGAGCGTACATCTGCAACAATGGAGG R: CCTCCATTGTTGCAGATGTACGCTCAACGTAAGGGTAGTC |
| C487A | F: GACTACCCTTACGTTGAGGCTACATCTGCAACAATGGAGG R: CCTCCATTGTTGCAGATGTAGCCTCAACGTAAGGGTAGTC |
| C565M | F: TGACTACCCTTACGTTGAGATGACATCTGCAACAATGGAG R: CTCCATTGTTGCAGATGTCATCTCAACGTAAGGGTAGTCA |
| C565R | F: TGACTACCCTTACGTTGAGCGTACATCTGCAACAATGGAG R: CTCCATTGTTGCAGATGTACGCTCAACGTAAGGGTAGTCA |
| C565A | F: TGACTACCCTTACGTTGAGGCTACATCTGCAACAATGGAG R: CTCCATTGTTGCAGATGTAGCCTCAACGTAAGGGTAGTCA |
| Y535W | F: CAATGGTGGTTTCGCATCTTGGGAATTAACAAGATCTTACCC R: GGGTAAGATCTTGTTAATTCCCAAGATGCGAAACCACCATTG |
| Y535L | F: CAATGGTGGTTTCGCATCTCTAGAATTAACAAGATCTTACCC R: GGGTAAGATCTTGTTAATTCTAGAGATGCGAAACCACCATTG |
| Y535A | F: CAATGGTGGTTTCGCATCTGCTGAATTAACAAGATCTTACCC R: GGGTAAGATCTTGTTAATTCAGCAGATGCGAAACCACCATTG |
| H260R | F: TCCAGGTAGAATGTGGTGTCGTTGTAGAATGGTTTATTTGCC R: GGCAAATAAACCATTCTACAACGACACCACATTCTACCTGGA |
| H260W | F: TCCAGGTAGAATGTGGTGTTGGTGTAGAATGGTTTATTTGCC R: GGCAAATAAACCATTCTACACCAACACCACATTCTACCTGGA |
| H260A | F: TCCAGGTAGAATGTGGTGTGCTTGTAGAATGGTTTATTTGCC R: GGCAAATAAACCATTCTACAAGCACACCACATTCTACCTGGA |
| DC486AA | F: AGATCACGGTTGGTTAATTTCTGCTGCTCTGCTGAAGGTTTAAAGGC R: GCCTTTAAACCTTCAGCAGAGCAGCAGAAATTAACCAACCGTGATCT |
| TAE4890AAA | F: CACGGTTGGTTAATTTCTGACTGTGCTGCTGCTGGTTTAAAGGCTGCTTTAATGTTA R: TAACATTAAAGCAGCCTTTAAACCAGCAGCAGCACAGTCAGAAATTAACCAACCGTG |
| QW113AA | F: GTCTTTTTATTCTTCTATCGCTACTTCTGATGGTAACGCTGCTTCTGATTTGGGTGGT R: ACCACCCAAATCAGAAGCAGCGTTACCATCAGAAGTAGCGATAGAAGAATAAAAAGAC |
| QW163AA | F: GTAGATATGTTTATAATCACGCTAACGAAGATGGTGGTGCTGGTTTGCATATTGAAGGT R: ACCTTCAATATGCAAACCAGCACCACCATCTTCGTTAGCGTGATTATAAACATATCTAC |
| QW603AA | F: GCAAATTTCTTAGAGAATATGGCTAGAACAGACGGTTCTGCTTACGGTTGTTGGGGTGTTT R: AAACACCCCAACAACCGTAAGCAGAACCGTCTGTTCTAGCCATATTCTCTAAGAAATTTGC |
| DCTAE4865A | F: ATCACGGTTGGTTAATTTCTGCTGCTGCTGCTGCTGGTTTAAAGGCTGCTTTAATG R: CATTAAAGCAGCCTTTAAACCAGCAGCAGCAGCAGCAGAAATTAACCAACCGTGAT |
| Site-saturation mutagenesis 260 | F: TCCAGGTAGAATGTGGTGTNNKTGTAGAATGGTTTATTT R: GGCAAATAAACCATTCTACAMNNACACCACATTCTACCT |















References
- Hill, R.A.; Connolly, J.D. Triterpenoids. Nat. Prod. Rep. 2013, 30, 1028–1065. [Google Scholar] [CrossRef] [PubMed]
- Nes, W.R. Role of sterols in membranes. Lipids 1974, 9, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Fazio, G.C.; Matsuda, S.P. On the origins of triterpenoid skeletal diversity. Phytochemistry 2004, 65, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Augustin, J.M.; Kuzina, V.; Andersen, S.B.; Bak, S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 2011, 72, 435–457. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Papadopoulou, K.K.; Osbourn, A. Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovganko, N.V.; Ananich, S.K. The Chemical Synthesis of Sterols: Latest Advances. Chem. Nat. Compd. 1999, 35, 229–259. [Google Scholar] [CrossRef]
- Yu, B.; Sun, J. Current synthesis of triterpene saponins. Chem. Asian J. 2010, 4, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Abe, I.; Rohmer, M.; Prestwich, G.D. Enzymatic Cyclization of Squalene and Oxidosqualene to Sterols and Triterpenes. Chem. Rev. 1993, 93, 2189–2206. [Google Scholar] [CrossRef]
- Kolesnikova, M.D.; Xiong, Q.; Lodeiro, S.; Hua, L.; Matsuda, S.P.T. Lanosterol biosynthesis in plants. Arch. Biochem. Biophys. 2006, 447, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Xiang, T.; Ohyama, K.; Seki, H.; Saito, K.; Muranaka, T.; Hayashi, H.; Katsube, Y.; Kushiro, T.; Shibuya, M. Lanosterol Synthase in Dicotyledonous Plants. Plant Cell Physiol. 2006, 47, 565–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eschenmoser, A.; Ruzicka, L.; Jeger, O.; Arigoni, D. Zur Kenntnis der Triterpene. 190. Mitteilung. Eine stereochemische Interpretation der biogenetischen Isoprenregel bei den Triterpenen. Helv. Chim. Acta 1955, 38, 1890–1904. [Google Scholar] [CrossRef]
- Wu, T.K.; Griffin, J.H. Conversion of a plant oxidosqualene-cycloartenol synthase to an oxidosqualene-lanosterol cyclase by random mutagenesis. Biochemistry 2002, 41, 8238–8244. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.K.; Liu, Y.T.; Chang, C.H.; Yu, M.T.; Wang, H.J. Site-saturated mutagenesis of histidine 234 of Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase demonstrates dual functions in cyclization and rearrangement reactions. J. Am. Chem. Soc. 2006, 128, 6414–6419. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Chen, Y.C.; Tseng, S.W.; Liu, Y.T.; Wen, H.Y.; Li, W.H.; Huang, C.Y.; Ko, C.Y.; Wang, T.T.; Wu, T.K. The cysteine 703 to isoleucine or histidine mutation of the oxidosqualene-lanosterol cyclase from Saccharomyces cerevisiae generates an iridal-type triterpenoid. Biochimie 2012, 94, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Wen, H.Y.; Shie, W.S.; Lu, C.T.; Li, M.E.; Liu, Y.T.; Li, W.H.; Wu, T.K. Protein engineering of oxidosqualene-lanosterol cyclase into triterpene monocyclase. Org. Biomol. Chem. 2013, 11, 4214–4219. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Hoshino, T. Catalytic function of the residues of phenylalanine and tyrosine conserved in squalene-hopene cyclases. Biosci. Biotechnol. Biochem. 2001, 65, 2233–2242. [Google Scholar] [CrossRef] [PubMed]
- Hart, E.A.; Ling, H.; Darr, L.B.; Wilson, W.K.; Jihai Pang, A.; Matsuda, S.P.T. Directed Evolution To Investigate Steric Control of Enzymatic Oxidosqualene Cyclization. An Isoleucine-to-Valine Mutation in Cycloartenol Synthase Allows Lanosterol and Parkeol Biosynthesis. J. Am. Chem. Soc. 1999, 121, 9887–9888. [Google Scholar] [CrossRef]
- Herrera, J.B.R.; And, W.K.W.; Matsuda, S.P.T. A Tyrosine-to-Threonine Mutation Converts Cycloartenol Synthase to an Oxidosqualene Cyclase that Forms Lanosterol as Its Major Product. J. Am. Chem. Soc. 2000, 122, 6765–6766. [Google Scholar] [CrossRef]
- Meyer, M.M.; Xu, R.; Matsuda, S.P. Directed evolution to generate cycloartenol synthase mutants that produce lanosterol. Org. Lett. 2002, 4, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Lodeiro, S.; Schulz-Gasch, T.; Matsuda, S. Enzyme Redesign: Two Mutations Cooperate to Convert Cycloartenol Synthase into an Accurate Lanosterol Synthase. J. Am. Chem. Soc. 2005, 127, 14132–14133. [Google Scholar] [CrossRef] [PubMed]
- Salmon, M.; Thimmappa, R.B.; Minto, R.E.; Melton, R.E.; Hughes, R.K.; O’Maille, P.E.; Hemmings, A.M.; Osbourn, A. A conserved amino acid residue critical for product and substrate specificity in plant triterpene synthases. Proc. Natl. Acad. Sci. USA 2016, 113, E4407–E4414. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Masukawa, Y.; Nakada, C.; Amari, K.; Nakano, C.; Hoshino, T. β-Amyrin synthase from Euphorbia tirucalli. Steric bulk, not the π-electrons of Phe, at position 474 has a key role in affording the correct folding of the substrate to complete the normal polycyclization cascade. Org. Biomol. Chem. 2014, 12, 3836–3846. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Liu, C.; Zhu, Y.; Zhang, J.; Men, Y.; Zeng, Y.; Sun, Y. Functional Characterization of Cucurbitadienol Synthase and Triterpene Glycosyltransferase Involved in Biosynthesis of Mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2015, 56, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Tang, Q.; Mo, C.; Bai, L.; Tu, D.; Ma, X. Cloning and characterization of squalene synthase and cycloartenol synthase from Siraitia grosvenorii. Acta Pharm. Sin. B 2017, 7, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M.; Adachi, S.; Ebizuka, Y. Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 2004, 35, 6995–7003. [Google Scholar] [CrossRef]
- Davidovich-Rikanati, R.; Shalev, L.; Baranes, N.; Meir, A.; Itkin, M.; Cohen, S.; Zimbler, K.; Portnoy, V.; Ebizuka, Y.; Shibuya, M. Recombinant yeast as a functional tool for understanding bitterness and cucurbitacin biosynthesis in watermelon (Citrullus spp.). Yeast 2015, 32, 103–114. [Google Scholar] [PubMed]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Thoma, R.; Schulzgasch, T.; D’Arcy, B.; Benz, J.; Aebi, J.; Dehmlow, H.; Hennig, M.; Stihle, M.; Ruf, A. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 2004, 432, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Poralla, K.; Hewelt, A.; Prestwich, G.D.; Abe, I.; Reipen, I.; Sprenger, G.A. specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem. Sci. 1994, 19, 157–158. [Google Scholar] [CrossRef]
- Corey, E.J.; Cheng, H.; Baker, C.H.; Matsuda, S.P.T.; Li, D.; Song, X. Studies on the Substrate Binding Segments and Catalytic Action of Lanosterol Synthase. Affinity Labeling with Carbocations Derived from Mechanism-Based Analogs of 2,3-Oxidosqualene and Site-Directed Mutagenesis Probes. J. Am. Chem. Soc. 1997, 119, 1289–1296. [Google Scholar] [CrossRef]
- Wu, T.K.; Liu, Y.T.; Chang, C.H. Histidine residue at position 234 of oxidosqualene-lanosterol cyclase from saccharomyces cerevisiae simultaneously influences cyclization, rearrangement, and deprotonation reactions. Chem. Biochem. 2005, 6, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.K. The evolutionary paths towards complexity: A metabolic perspective. New Phytol. 2014, 201, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Banta, A.B.; Wei, J.H.; Gill, C.C.; Giner, J.L.; Welander, P.V. Synthesis of arborane triterpenols by a bacterial oxidosqualene cyclase. Proc. Natl. Acad. Sci. USA 2016, 114, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Hashimoto, I.; Masukawa, Y.; Hoshino, T. Effect of cation-π interactions and steric bulk on the catalytic action of oxidosqualene cyclase: A case study of Phe728 of β-amyrin synthase from Euphorbia tirucalli L. Chem. Eur. J. 2013, 19, 17150–17158. [Google Scholar] [CrossRef] [PubMed]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene Biosynthesis in Plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef] [PubMed]
- Wendt, K.U.; Lenhart, A.; Schulz, G.E. The structure of the membrane protein squalene-hopene cyclase at 2.0 å resolution 1. J. Mol. Biol. 1999, 286, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Shibuya, M.; Masuda, K.; Ebizuka, Y. Mutational Studies on Triterpene Synthases: Engineering Lupeol Synthase into β-Amyrin Synthase. J. Am. Chem. Soc. 2000, 122, 6818–6824. [Google Scholar] [CrossRef]
- Schulz–Gasch, T.; Stahl, M. Mechanistic insights into oxidosqualene cyclizations through homology modeling. J. Comput. Chem. 2003, 24, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.; Miller, S.M.; Lai, M.H.; Kirsch, D.R. Cloning and characterization of the 2,3-oxidosqualene cyclase-coding gene of Candida albicans. Gene 1990, 87, 177–183. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, Y.; Ovchinnikov, S.; Greisen, P., Jr.; Huang, S.; Yi, S. New insights into substrate folding preference of plant OSCs. Sci. Bull. 2016, 61, 1407–1412. [Google Scholar] [CrossRef]
- Vickers, C.E.; Bydder, S.F.; Zhou, Y.; Nielsen, L.K. Dual gene expression cassette vectors with antibiotic selection markers for engineering in Saccharomyces cerevisiae. Microb. Cell Fact. 2013, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 2015, 19, 1639–1662. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2010, 28, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Geuflores, F.; Noureldin, H.H.; Nielsen, M.T.; Halkier, B.A. USER fusion: A rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 2007, 35, e55. [Google Scholar] [CrossRef] [PubMed]






| Amino Acids Substitution | Product Profile | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Ala (A) | 18 | 19 | 5 | 37 | 5 | 16 | |
| Cys (C) | 43 | 24 | 32 | ||||
| Asp (D) | 20 | 59 | 21 | ||||
| Glu (E) | 43 | 34 | 23 | ||||
| Phe (F) | 53 | 47 | |||||
| Gly (G) | 10 | 18 | 52 | 20 | |||
| Ile (I) | 24 | 36 | 23 | 17 | |||
| Lys (K) | 58 | 9 | 33 | ||||
| Leu (L) | 11 | 32 | 31 | 4 | 21 | ||
| Met (M) | 43 | 29 | 17 | 11 | |||
| Asn (N) | 48 | 9 | 32 | 2 | 8 | ||
| Pro (P) | 15 | 21 | 22 | 18 | 24 | ||
| Gln (Q) | 71 | 3 | 17 | 9 | |||
| Arg (R) | 20 | 6 | 49 | 4 | 22 | ||
| Ser (S) | 8 | 56 | 36 | ||||
| Thr (T) | 10 | 55 | 35 | ||||
| Val (V) | 25 | 40 | 13 | 22 | |||
| Trp (W) | 46 | 24 | 30 | ||||
| Tyr (Y) | 20 | 52 | 28 | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, J.; Liu, J.; Liao, J.; Luo, Z.; Ma, X.; Ma, G. Identification of Key Amino Acid Residues Determining Product Specificity of 2,3-Oxidosqualene Cyclase in Siraitia grosvenorii. Catalysts 2018, 8, 577. https://doi.org/10.3390/catal8120577
Qiao J, Liu J, Liao J, Luo Z, Ma X, Ma G. Identification of Key Amino Acid Residues Determining Product Specificity of 2,3-Oxidosqualene Cyclase in Siraitia grosvenorii. Catalysts. 2018; 8(12):577. https://doi.org/10.3390/catal8120577
Chicago/Turabian StyleQiao, Jing, Jiushi Liu, Jingjing Liao, Zuliang Luo, Xiaojun Ma, and Guoxu Ma. 2018. "Identification of Key Amino Acid Residues Determining Product Specificity of 2,3-Oxidosqualene Cyclase in Siraitia grosvenorii" Catalysts 8, no. 12: 577. https://doi.org/10.3390/catal8120577