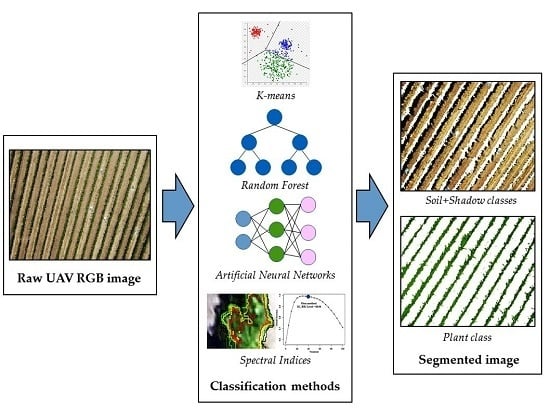
Detection and Segmentation of Vine Canopy in Ultra-High Spatial Resolution RGB Imagery Obtained from Unmanned Aerial Vehicle (UAV): A Case Study in a Commercial Vineyard
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. UAV Imagery Acquisition
2.3. Description of the Classification Models
2.3.1. Spectral Indices as Classification Methods
2.3.2. K-Means Clustering Method
2.3.3. Artificial Neural Networks
2.3.4. Random Forest
2.4. Accuracy Assessment
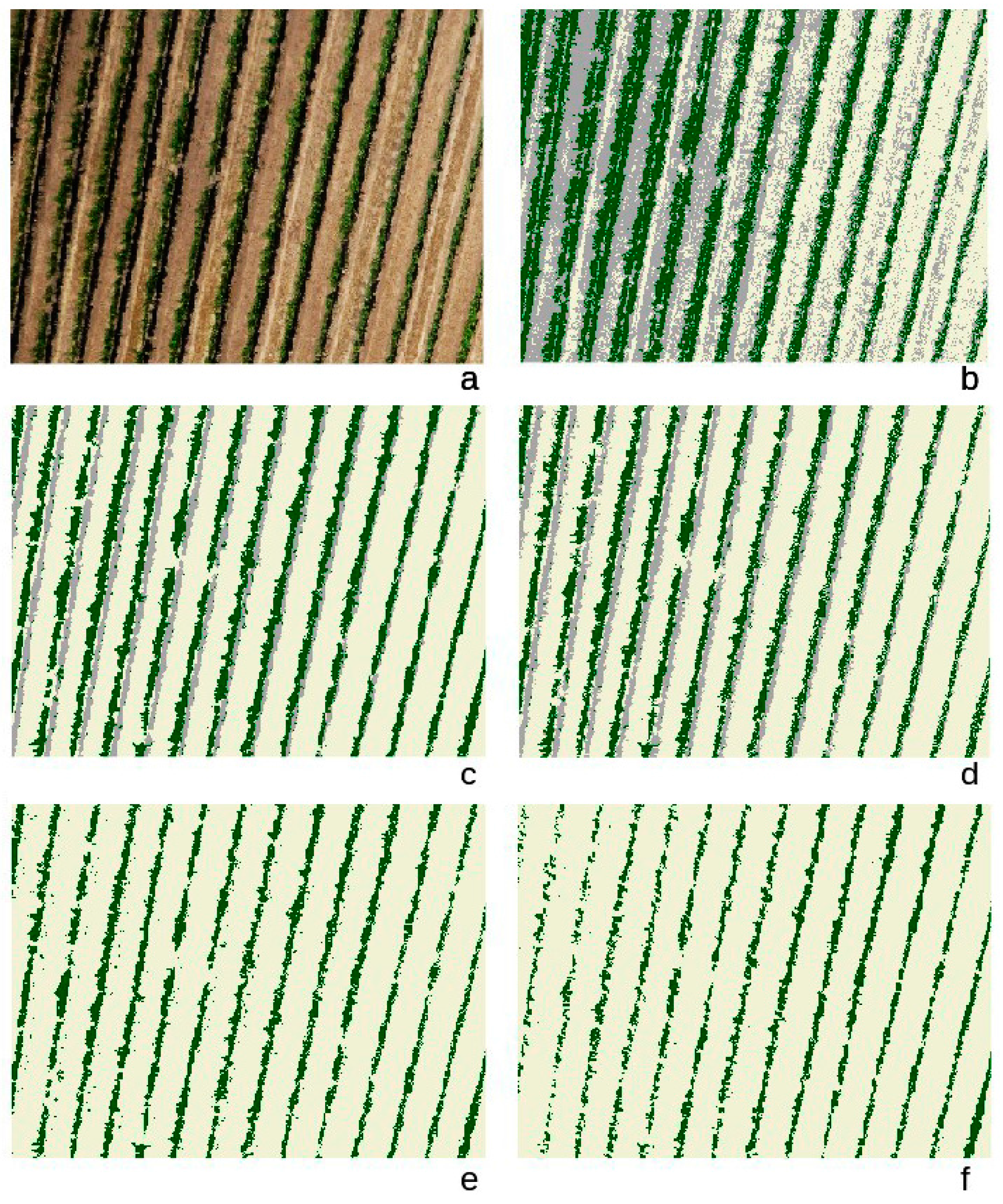
3. Results
4. Discussion
4.1. Perspectives and General Study Limitations
4.2. Accuracy of Classification Methods
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Proffitt, T.; Bramley, R.; Lamb, D.; Winter, E. Precision Viticulture: A New Era in Vineyard Management and Wine Production; Winetitles: Ashford, South Australia, 2006. [Google Scholar]
- Comba, L.; Gay, P.; Primicerio, J.; Aimonino, D.R. Vineyard detection from unmanned aerial systems images. Comput. Electron. Agric. 2015, 114, 78–87. [Google Scholar] [CrossRef]
- Bramley, R.G.V.; Lamb, D.W. Making sense of vineyard variability in Australia. Available online: http://www.cse.csiro.au/client_serv/resources/Bramley_Chile_Paper_h.pdf (accessed on 13 March 2017).
- Gutierrez-Rodriguez, M.; Escalante-Estrada, J.A.; Rodriguez-Gonzalez, M.T. Canopy Reflectance, Stomatal Conductance, and Yield of Phaseolus vulgaris L. and Phaseolus coccinues L. Under Saline Field Conditions. Int. J. Agric. Biol. 2005, 7, 491–494. [Google Scholar]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309–317. [Google Scholar]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Bachmann, F.; Herbst, R.; Gebbers, R.; Hafner, V.V. Micro UAV based georeferenced orthophoto generation in VIS + NIR for precision agriculture. Available online: http://www.int-arch-photogramm-remote-sens-spatial-inf-sci.net/XL-1-W2/11/2013/isprsarchives-XL-1-W2-11-2013.pdf (accessed on 13 March 2017).
- Smit, J.L.; Sithole, G.; Strever, A.E. Vine signal extraction—An application of remote sensing in precision viticulture. S. Afr. J. Enol. Vitic. 2010, 31, 65–74. [Google Scholar] [CrossRef]
- Hall, A.; Louis, J.; Lamb, D. Characterising and mapping vineyard canopy using high-spatial-resolution aerial multispectral images. Comput. Geosci. 2003, 29, 813–822. [Google Scholar] [CrossRef]
- Nebiker, S.; Annen, A.; Scherrer, M.; Oesch, D. A light-weight multispectral sensor for micro UAV—Opportunities for very high resolution airborne remote sensing. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, XXXVII, 1193–1200. [Google Scholar]
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, J.; Gong, J. UAV Remote Sensing for Urban Vegetation Mapping Using Random Forest and Texture Analysis. Remote Sens. 2015, 7, 1074–1094. [Google Scholar] [CrossRef]
- Candiago, S.; Remondino, F.; De Giglio, M.; Dubbini, M.; Gattelli, M. Evaluating Multispectral Images and Vegetation Indices for Precision Farming Applications from UAV Images. Remote Sens. 2015, 7, 4026–4047. [Google Scholar] [CrossRef]
- Mathews, A.J.; Jensen, J.L.R. Visualizing and quantifying vineyard canopy LAI using an unmanned aerial vehicle (UAV) collected high density structure from motion point cloud. Remote Sens. 2013, 5, 2164–2183. [Google Scholar] [CrossRef]
- Kalisperakis, I.; Stentoumis, C.; Grammatikopoulos, L.; Karantzalos, K. Leaf area index estimation in vineyards from UAV hyperspectral data, 2D image mosaics and 3D canopy surface models. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci.-ISPRS Arch. 2015, 40, 299–303. [Google Scholar] [CrossRef]
- Rabatel, G.; Delenne, C.; Deshayes, M. A non-supervised approach using Gabor filters for vine-plot detection in aerial images. Comput. Electron. Agric. 2008, 62, 159–168. [Google Scholar] [CrossRef]
- Nolan, A.P.; Park, S.; Fuentes, S.; Ryu, D.; Chung, H. Automated detection and segmentation of vine rows using high resolution UAS imagery in a commercial vineyard. In Proceedings of the 21st International Congress on Modelling and Simulation, Gold Coast, Australia, 29 November–4 December 2015; pp. 1406–1412.
- Ranchin, T.; Naert, B.; Albuisson, M.; Boyer, G.; Astrand, P. An automatic method for vine detection in airborne imagery using wavelet transform and multiresolution analysis. Photogramm. Eng. Remote Sens. 2001, 67, 91–98. [Google Scholar]
- Wassenaar, T.; Robbez-Masson, J.; Andrieux, P.; Baret, F. Vineyard identification and description of spatial crop structure by per-field frequency analysis. Int. J. Remote Sens. 2002, 23, 3311–3325. [Google Scholar] [CrossRef]
- Puletti, N.; Perria, R.; Storchi, P. Unsupervised classification of very high remotely sensed images for grapevine rows detection. Eur. J. Remote Sens. 2014, 47, 45–54. [Google Scholar] [CrossRef]
- Richardson, A.D.; Jenkins, J.P.; Braswell, B.H.; Hollinger, D.Y.; Ollinger, S.V.; Smith, M.L. Use of digital webcam images to track spring green-up in a deciduous broadleaf forest. Oecologia 2007, 152, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Liao, P.S.; Chen, T.S.; Chung, P.C. A fast algorithm for multilevel thresholding. J. Inf. Sci. Eng. 2001, 17, 713–727. [Google Scholar]
- Huang, D.Y.; Lin, T.W.; Hu, W.C. Automatic multilevel thresholding based on two-stage Otsu’s method with cluster determination by valley estimation. Int. J. Innov. Comput. Inf. Control 2011, 7, 5631–5644. [Google Scholar]
- Hunag, M.-C.; Wu, J.; Cang, J.; Yang, D. An Efficient k-Means Clustering Algorithm Using Simple Partitioning. J. Inf. Sci. Eng. 2005, 1177, 1157–1177. [Google Scholar]
- R Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2015. Available online: https://www.R-project.org/ (accessed on 15 March 2017).
- Lloyd, S.P. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 128–137. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Shapire, R.; Freund, Y.; Bartlett, P.; Lee, W.S. Boosting the Margin: A New Explanation for the Effectiveness of Voting Methods. Ann. Stat. 1998, 26, 1651–1686. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random Forests for Classification in Ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Liaw, A.; Wiener, M. Classification and Regression by Random Forest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Baluja, J.; Diago, M.P.; Balda, P.; Zorer, R.; Meggio, F.; Morales, F.; Tardaguila, J. Assessment of vineyard water status variability by thermal and multispectral imagery using an unmanned aerial vehicle (UAV). Irrig. Sci. 2012, 30, 511–522. [Google Scholar] [CrossRef]
- Gevrey, M.; Dimopoulos, I.; Lek, S. Review and comparison of methods to study the contribution of variables in artificial neural network models. Ecol. Modell. 2003, 160, 249–264. [Google Scholar] [CrossRef]
- Mathews, A.J. A Practical UAV Remote Sensing Methodology to Generate Multispectral Orthophotos for Vineyards: Estimation of Spectral Reflectance. Int. J. Appl. Geospatial Res. 2015, 6, 65–87. [Google Scholar] [CrossRef]
- Delenne, C.; Durrieu, S.; Rabatel, G.; Deshayes, M. From pixel to vine parcel: A complete methodology for vineyard delineation and characterization using remote-sensing data. Comput. Electron. Agric. 2010, 70, 78–83. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef]
- Karakizi, C.; Oikonomou, M.; Karantzalos, K. Vineyard detection and vine variety discrimination from very high resolution satellite data. Remote Sens. 2016, 8, 1–25. [Google Scholar] [CrossRef]

| Characteristic | Description |
|---|---|
| Type | Quadcopter |
| Dimensions | Diameter 100 cm, height 45 cm |
| Weight | 5.4 kg with batteries (maximum weight on fly 9.0 kg) |
| Engine power | 4 multirotor motors × 250 W gearless brushless motors powered by a 14.8 V battery |
| Auto pilot | HKPilot Mega 2.7 |
| Material | Carbon with delrin inserts |
| Payload | Approximately, 3.0 kg |
| Flight mode | Automatic with waypoint or based on radio control |
| Endurance | Approximately, 21 min (hovering flight time) and 18 min (acquisition flight time) |
| Ground Control Station | 8-channels, UHF modem, telemetry for real-time flight control |
| Onboard imaging sensor | Conventional RGB camera |
| Method | Predictors | Training Samples | Parameters |
|---|---|---|---|
| K-means | R, G, B | - | 3 centers, 50 max iterations |
| K-means.ex | R, G, B, G%, 2G_RBi | - | 3 centers, 50 max iterations |
| ANN | R, G, B | 672 | size = 4, decay = 0.1 |
| ANN.ex | R, G, B, G%, 2G_RBi | 672 | size = 5, decay = 0.1 |
| RForest | R, G, B | 672 | trees = 500 |
| RForest.ex | R, G, B, G%, 2G_RBi | 672 | trees = 500 |
| Method | Flight1 DOY315 | Flight2 DOY22 | Flight3 DOY29 | Flight4 DOY63 | Flight5 DOY72 | Flight6 DOY78 | **Avg. |
|---|---|---|---|---|---|---|---|
| Overall Accuracy (Kappa Index) | |||||||
| * G% | 0.98 (0.98) | 0.96 (0.91) | 0.98 (0.97) | 0.94 (0.88) | 0.97 (0.93) | 0.93 (0.85) | 0.96 (0.92) |
| * 2G_RBi | 0.99 (0.98) | 0.97 (0.94) | 0.99 (0.98) | 0.97 (0.94) | 0.97 (0.94) | 0.98 (0.95) | 0.98 (0.96) |
| K-means | 0.81 (0.71) | 0.58 (0.35) | 0.53 (0.29) | 0.64 (0.47) | 0.54 (0.29) | 0.48 (0.21) | 0.60 (0.39) |
| K-means.ex | 0.96 (0.83) | 0.60 (0.38) | 0.55 (0.32) | 0.71 (0.57) | 0.56 (0.32) | 0.49 (0.22) | 0.64 (0.46) |
| ANN | 0.98 (0.97) | 0.96 (0.93) | 0.99 (0.98) | 0.96 (0.94) | 0.98 (0.97) | 0.93 (0.89) | 0.90 (0.95) |
| ANN.ex | 0.97 (0.96) | 0.96 (0.94) | 0.99 (0.99) | 0.96 (0.94) | 0.98 (0.98) | 0.94 (0.90) | 0.97 (0.95) |
| RForest | 0.96 (0.94) | 0.90 (0.84) | 0.88 (0.82) | 0.83 (0.73) | 0.90 (0.84) | 0.75 (0.60) | 0.87 (0.79) |
| RForest.ex | 0.97 (0.96) | 0.96 (0.94) | 0.98 (0.96) | 0.91 (0.86) | 0.95 (0.93) | 0.89 (0.83) | 0.94 (0.91) |
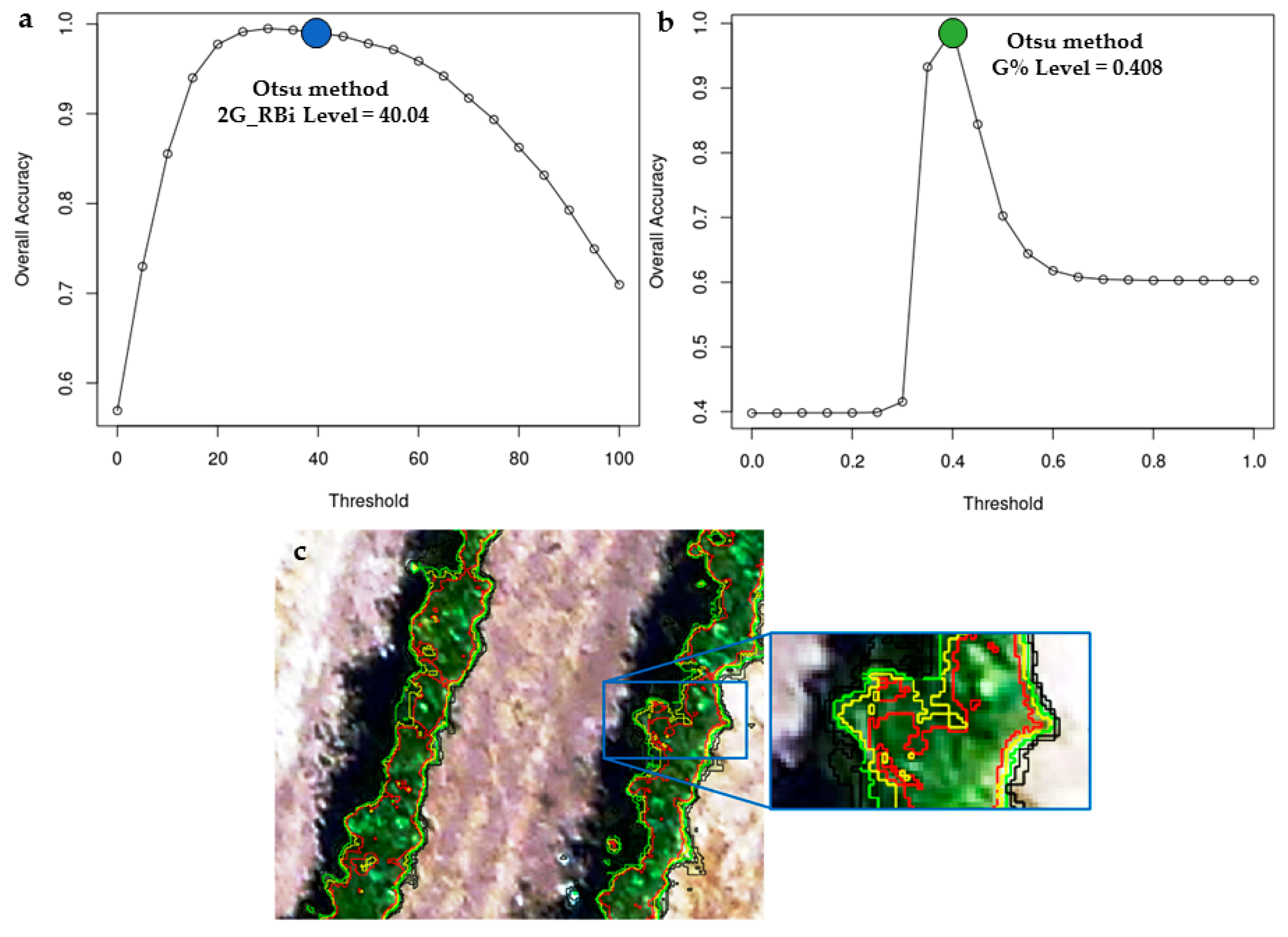
| Threshold Values Estimated Using the Otsu Method | |||||||
| * G% | 0.45 | 0.40 | 0.41 | 0.39 | 0.39 | 0.39 | 0.40 |
| * 2G_RBi | 63.27 | 37.22 | 40.04 | 34.41 | 33.28 | 31.08 | 39.88 |
| Sensitivity (Plant Class) | |||||||
| K-means | 0.02 | 0.00 | 0.55 | 0.42 | 0.07 | 0.56 | 0.27 |
| K-means.ex | 0.00 | 0.00 | 0.38 | 0.46 | 0.23 | 0.39 | 0.24 |
| ANN | 1.00 | 0.93 | 1.00 | 0.92 | 0.95 | 0.84 | 0.94 |
| ANN.ex | 1.00 | 0.94 | 1.00 | 0.92 | 0.96 | 0.86 | 0.95 |
| RForest | 0.94 | 0.85 | 0.81 | 0.65 | 0.82 | 0.67 | 0.79 |
| RForest.ex | 0.98 | 0.92 | 0.95 | 0.78 | 0.88 | 0.73 | 0.87 |
| Sensitivity (Shadow Class) | |||||||
| K-means | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.17 |
| K-means.ex | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.33 |
| ANN | 0.94 | 0.94 | 0.96 | 0.98 | 1.00 | 0.98 | 0.97 |
| ANN.ex | 0.92 | 0.94 | 0.97 | 0.96 | 1.00 | 0.98 | 0.96 |
| RForest | 0.94 | 0.78 | 0.80 | 0.84 | 0.86 | 0.42 | 0.77 |
| RForest.ex | 0.94 | 0.96 | 0.97 | 1.00 | 1.00 | 1.00 | 0.98 |
| Sensitivity (Soil Class) | |||||||
| K-means | 0.00 | 0.00 | 0.61 | 0.68 | 0.01 | 0.64 | 0.32 |
| K-means.ex | 0.01 | 0.00 | 0.62 | 0.78 | 0.00 | 0.00 | 0.23 |
| ANN | 0.98 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 |
| ANN.ex | 0.97 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 |
| RForest | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 1.00 |
| RForest.ex | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Input Variable | ANN | ANN.ex | RForest | RForest.ex |
|---|---|---|---|---|
| R | 100 | 52.94 | 66.86 | 55.45 |
| G | 73.21 | 0 | 100 | 0 |
| B | 0 | 45.31 | 0 | 16.77 |
| G% | - | 100 | - | 100 |
| 2G_RBi | - | 50.12 | - | 82.96 |
| Method | Input Data | Spatial Resolution | Best Results from the Research Study | Reference |
|---|---|---|---|---|
| Dynamic segmentation, Hough Space Clustering and Total Least Squares techniques | UAV. Near Infrared images | 5.6 cm ground resolution | Average percentage of correctly detected vine-rows 95.13% | [2] |
| Histogram filtering, Contour recognition, and Skeletonisation process | UAV. Near Infrared images. | 4.0 cm ground resolution | Average precision 0.971. Sensitivity 0.971. | [18] |
| Object-based procedure and Ward’s Modified Method | Aircraft. RGB images. | 30 cm ground resolution | OA for both methods 0.87 | [21] |
| Object-based procedure | Satellite. Multispectral WorldView-2 images. | 50 cm Panchromatic imagery. 200 cm multispectral imagery. | OA values above 96% for all datasets | [41] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poblete-Echeverría, C.; Olmedo, G.F.; Ingram, B.; Bardeen, M. Detection and Segmentation of Vine Canopy in Ultra-High Spatial Resolution RGB Imagery Obtained from Unmanned Aerial Vehicle (UAV): A Case Study in a Commercial Vineyard. Remote Sens. 2017, 9, 268. https://doi.org/10.3390/rs9030268
Poblete-Echeverría C, Olmedo GF, Ingram B, Bardeen M. Detection and Segmentation of Vine Canopy in Ultra-High Spatial Resolution RGB Imagery Obtained from Unmanned Aerial Vehicle (UAV): A Case Study in a Commercial Vineyard. Remote Sensing. 2017; 9(3):268. https://doi.org/10.3390/rs9030268
Chicago/Turabian StylePoblete-Echeverría, Carlos, Guillermo Federico Olmedo, Ben Ingram, and Matthew Bardeen. 2017. "Detection and Segmentation of Vine Canopy in Ultra-High Spatial Resolution RGB Imagery Obtained from Unmanned Aerial Vehicle (UAV): A Case Study in a Commercial Vineyard" Remote Sensing 9, no. 3: 268. https://doi.org/10.3390/rs9030268
APA StylePoblete-Echeverría, C., Olmedo, G. F., Ingram, B., & Bardeen, M. (2017). Detection and Segmentation of Vine Canopy in Ultra-High Spatial Resolution RGB Imagery Obtained from Unmanned Aerial Vehicle (UAV): A Case Study in a Commercial Vineyard. Remote Sensing, 9(3), 268. https://doi.org/10.3390/rs9030268