Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection
Abstract
1. Introduction
1.1. Phenology-Based Species Classification
1.2. Objectives
- Many studies focus on optimal small study sites with a limited number of carefully chosen species and individuals. The sites do not necessarily represent the heterogeneity of the local habitat (species richness and inter-species variability), and therefore the findings are of limited contribution to applicative mapping over larger areas or different regions.
- Most studies follow a data-driven approach and focus on maximizing total classification accuracy, without referring to the fundamental factors that affect the classification success on the species level.
- In general, cost-effectiveness does not appear to be a major consideration. This is significant, since many studies use state-of-the-art sensors that are not accessible to most practitioners, and their implementation over large areas is limited.
- Phenology patterns are a key component in the discrimination of vegetation species, and near-surface sensors can be a reliable tool for obtaining a full annual time-series of individual plants.
- The extraction of optimal dates for classification (when and how many) from a full near-surface time series can assist in optimizing the classification accuracy of sequential overhead data acquisition.
- A large and representative sample of individuals reflects the variability within and between species, and is therefore essential for obtaining robust insights that can enable future implementation in similar areas.
1.3. Classification of East Mediterranean Vegetation by Remote Sensing
2. Materials and Methods
2.1. Near-Surface Data Collection and Preprocessing
2.2. Selecting Optimal Acquisition Times for Species Classification on the Basis of the Near-Surface Time Series
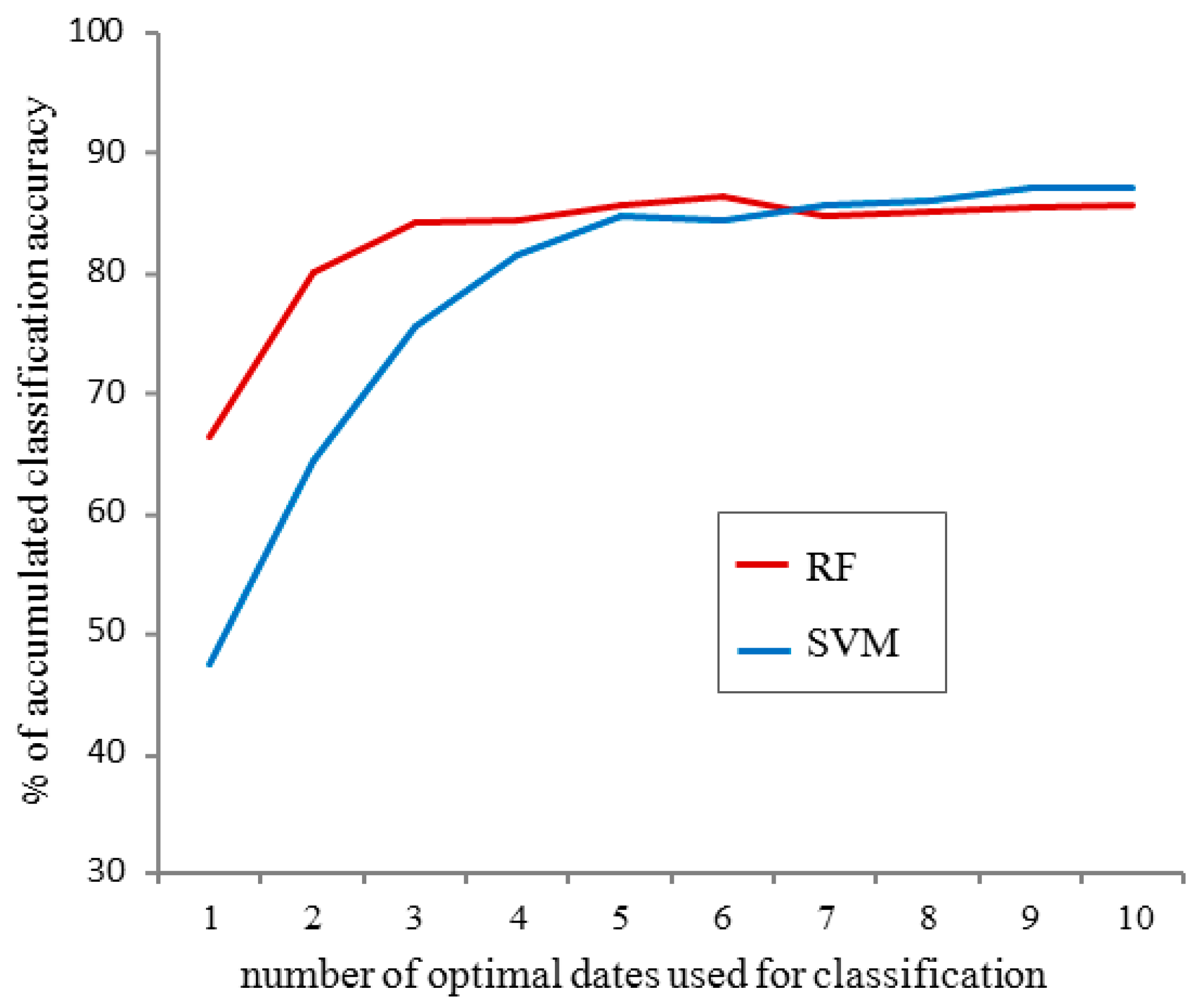
- Comparison of the accumulated classification accuracy for the first ten optimal dates, using separate runs for testing the use of the RF and SVM classifier. We found that RF led to better classification accuracy compared to SVM, and therefore the following scenarios were all run using RF only (see Results). This was done using all spectral indices for observations in the S’S site only.
- Comparison of the accumulated classification accuracy for the first ten optimal dates in all four sites, using RF, and all spectral indices were used as input.
- Examining the accumulated classification accuracy for the first ten optimal dates, comparing between using different combinations of the spectral indices (see Results), in order to examine their contribution to classification accuracy. This was done by using RF, for observations in the S’S site only.
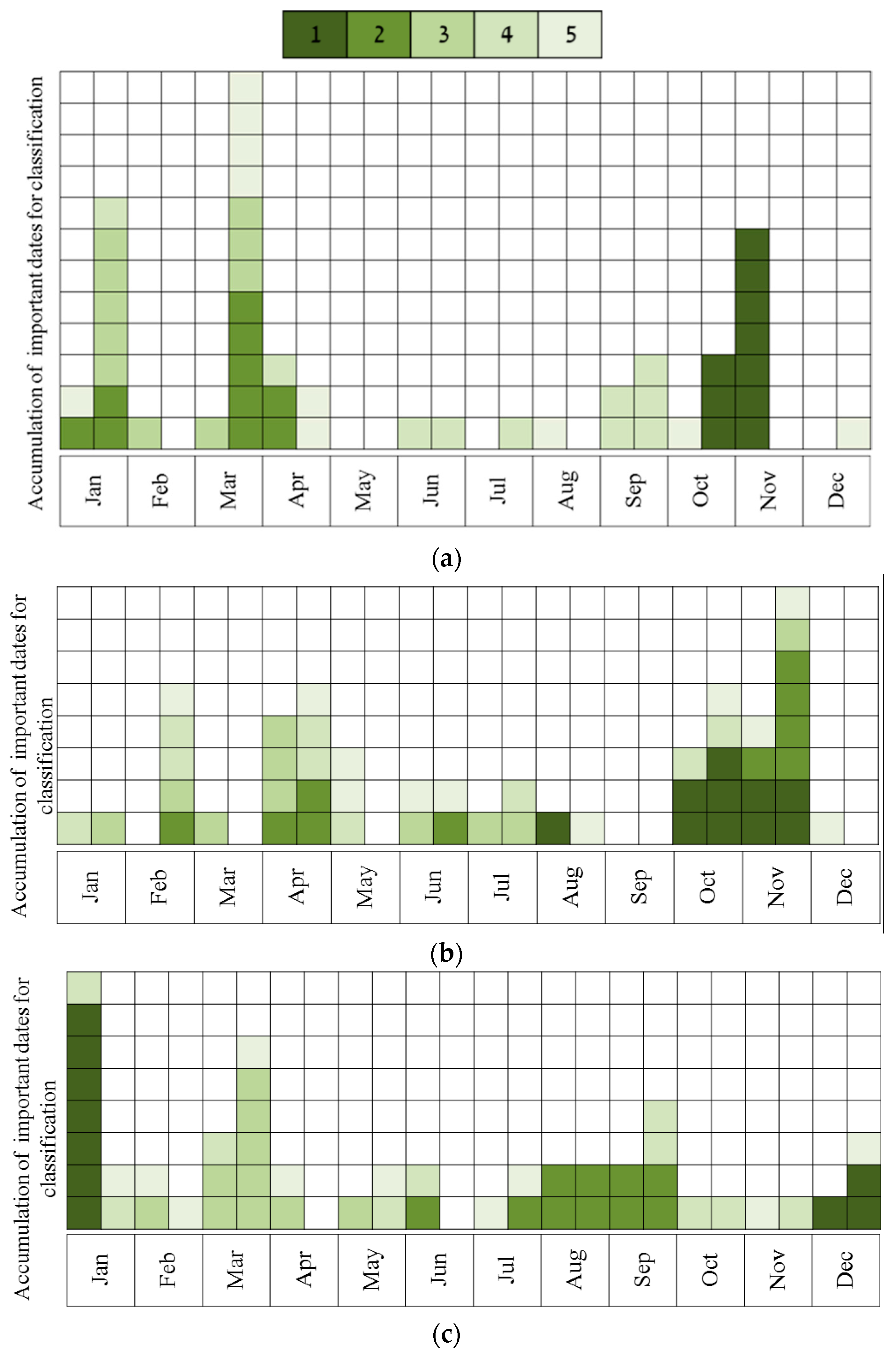
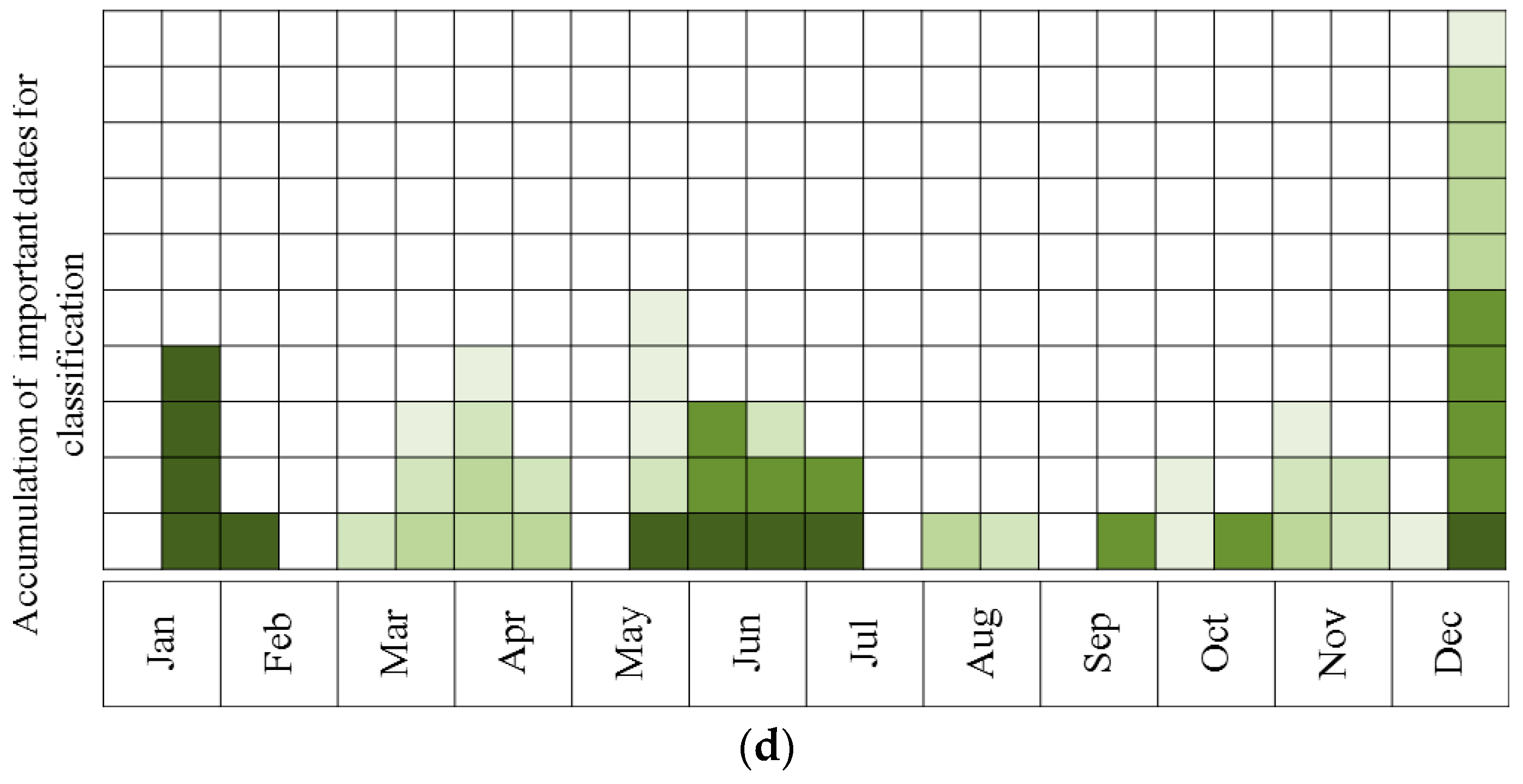
- Describing the results of ten sequential runs—the most important five dates for classification of all species (the number of UAV acquisition times was set to five due to the findings of the above tests, see Results). This was carried out for all four sites. Ten sequential runs were used because of the need to evaluate the robustness of the results, since the classification process included random components (RF and the internal division of subsets for cross-validation). This was done by using RF, and all spectral indices as input.
- Describing the results of ten sequential runs—the most important five dates for discrimination between Pinus halepensis (evergreen conifer) and Quercus calliprinos (evergreen broad-leaved) in the S’S site. The classification was carried out by RF, using all spectral indices. The purpose of this scenario was to examine the proposed methodology for the discrimination of specific species. These species were selected because of the practical management need for mapping P. halepensis, which is a dominant factor in the occurrence of intensive forest fires in Israel, and which is additionally spreading from dense plantations to the surrounding natural vegetation, dominated by Q. calliprinos [93,94].
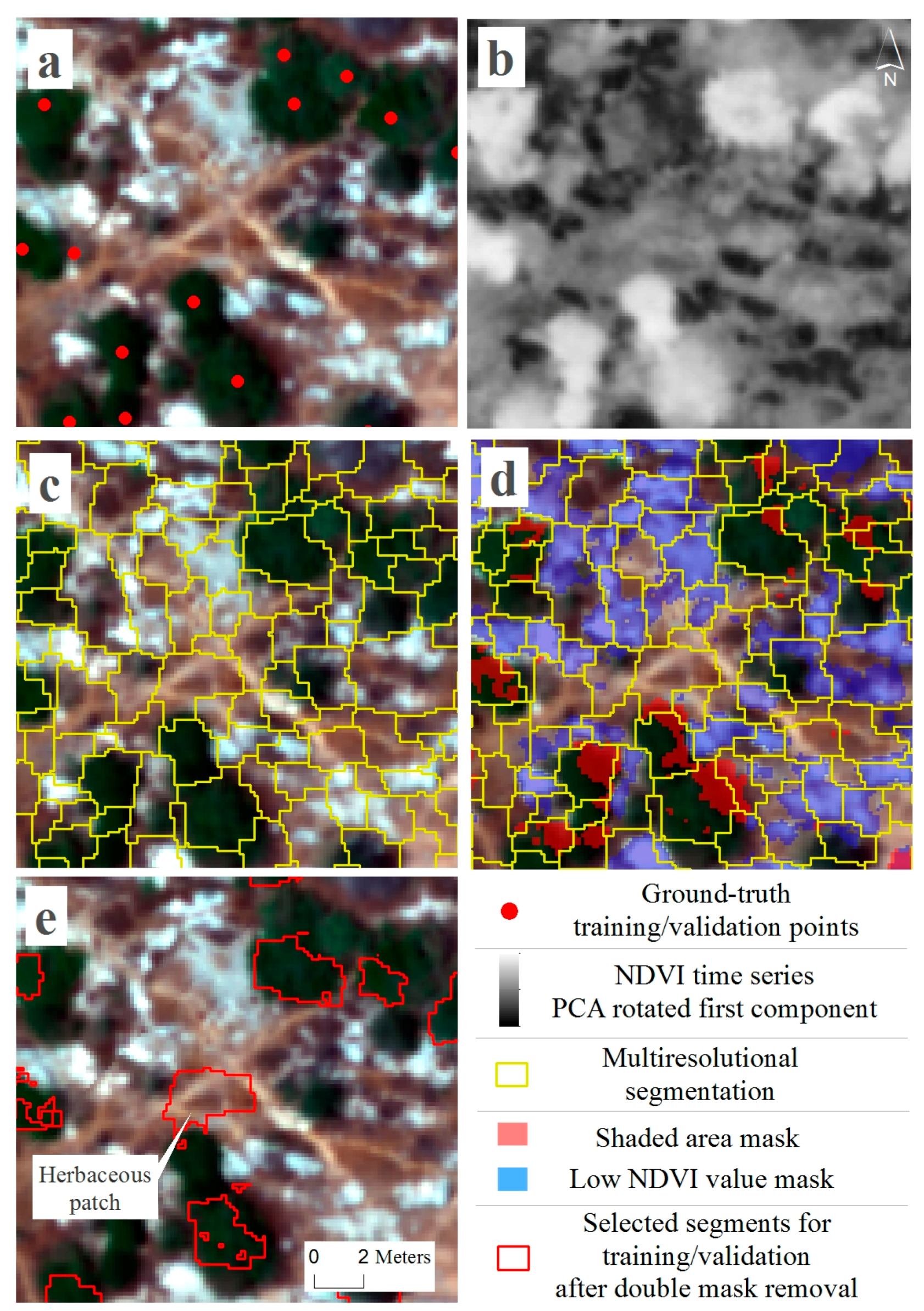
2.3. Overhead Data Acquisition and Species Classification
3. Results
3.1. Obtaining Optimal Dates for Species Classification from Near-Surface Observations
3.2. Overhead Data Acquisition and Species Classification
4. Discussion
4.1. Species-Driven Approach as a Key Component for Applicative Species Mapping by Remote Sensing
4.2. Near-Surface Phenological Observations and Sequential UAV Repetitive Imagery as a Cost-Effective Methodology for Detailed Vegetation Mapping
4.3. Relevant Implications for Satellite Imagery
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Kerr, J.T.; Ostrovsky, M. From space to species: Ecological applications for remote sensing. Trends Ecol. Evol. 2003, 18, 299–305. [Google Scholar] [CrossRef]
- Mulla, D.J. Twenty five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2003, 114, 358–371. [Google Scholar] [CrossRef]
- Rocchini, D.; Boyd, D.S.; Féret, J.B.; Foody, G.M.; He, K.S.; Lausch, A.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecol. Conserv. 2015, 2, 25–36. [Google Scholar] [CrossRef]
- Vilà, M.; Vayreda, J.; Comas, L.; Ibáñez, J.J.; Mata, T.; Obón, B. Species richness and wood production: A positive association in Mediterranean forests. Ecol. Lett. 2007, 10, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Müllerová, J.; Bruna, J.; Dvorák, P.; Bartalos, T.; Vítková, M. Does the Data Resolution/origin Matter? Satellite, Airborne and UAV Imagery to Tackle Plant Invasions. Int. Arch. Photogram. Remote Sens. Spat. Inf. Sci. 2016, 41, 903–908. [Google Scholar] [CrossRef]
- Roth, K.L.; Roberts, D.A.; Dennison, P.E.; Peterson, S.H.; Alonzo, M. The impact of spatial resolution on the classification of plant species and functional types within imaging spectrometer data. Remote Sens. Environ. 2015, 171, 45–57. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Mapping tree species in tropical seasonal semi-deciduous forests with hyperspectral and multispectral data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (RandomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Paz-Kagan, T.; Caras, T.; Herrmann, I.; Shachak, M.; Karnieli, A. Multiscale Mapping of Species Diversity under Changed Land-Use Using Imaging Spectroscopy. Ecol. Appl. 2017. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2008, 7, 269–276. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and LIDAR remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef]
- Kozhoridze, G.; Orlovsky, N.; Orlovsky, L.; Blumberg, D.G.; Golan-Goldhirsh, A. Remote sensing models of structure-related biochemicals and pigments for classification of trees. Remote Sens. Environ. 2016, 186, 184–195. [Google Scholar] [CrossRef]
- Galidaki, G.; Gitas, I. Mediterranean forest species mapping using classification of Hyperion imagery. Geocarto Int. 2015, 30, 48–61. [Google Scholar] [CrossRef]
- Lieth, H. Phenology in productivity studies. In Analysis of Temperate Forest Ecosystems; Springer: Berlin/Heidelberg, Germany, 1973; pp. 29–46. [Google Scholar]
- Morisette, J.T.; Richardson, A.D.; Knapp, A.K.; Fisher, J.I.; Graham, E.A.; Abatzoglou, J.; Wilson, B.; Breshears, D.; Henebry, G.; Hanes, J.; et al. Tracking the rhythm of the seasons in the face of global change: Phenological research in the 21st century. Front. Ecol. Environ. 2009, 7, 253–260. [Google Scholar] [CrossRef]
- Rathcke, B.; Lacey, E.P. Phenological patterns of terrestrial plants. Annu. Rev. Ecol. Evol. 1985, 16, 179–214. [Google Scholar] [CrossRef]
- Dudley, K.L.; Dennison, P.E.; Roth, K.L.; Roberts, D.A.; Coates, A.R. A multi-temporal spectral library approach for mapping vegetation species across spatial and temporal phenological gradients. Remote Sens. Environ. 2015, 167, 121–134. [Google Scholar] [CrossRef]
- Hill, R.A.; Wilson, A.K.; George, M.; Hinsley, S.A. Mapping tree species in temperate deciduous woodland using time-series multi-spectral data. Appl. Veg. Sci. 2010, 13, 86–99. [Google Scholar] [CrossRef]
- Key, T.; Warner, T.A.; McGraw, J.B.; Fajvan, M.A. A comparison of multispectral and multitemporal information in high spatial resolution imagery for classification of individual tree species in a temperate hardwood forest. Remote Sens. Environ. 2001, 75, 100–112. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Mathieu, R.; Mutanga, O.; Ramoelo, A.; Kaszta, Ż.; Wolff, E. Multi-phenology WorldView-2 imagery improves remote sensing of savannah tree species. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 65–73. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G.P. Multi-temporal hyperspectral mixture analysis and feature selection for invasive species mapping in rainforests. Remote Sens. Environ. 2013, 136, 14–27. [Google Scholar] [CrossRef]
- Anderson, K.; Gaston, K.J. Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Front. Ecol. Environ. 2013, 11, 138–146. [Google Scholar] [CrossRef]
- Kaneko, K.; Nohara, S. Review of effective vegetation mapping using the UAV (Unmanned Aerial Vehicle) method. IJGIS 2014, 6, 733. [Google Scholar] [CrossRef]
- Pajares, G. Overview and current status of remote sensing applications based on unmanned aerial vehicles (UAVs). Photogramm. Eng. Remote Sens. 2015, 81, 281–329. [Google Scholar] [CrossRef]
- Watts, A.C.; Ambrosia, V.G.; Hinkley, E.A. Unmanned aircraft systems in remote sensing and scientific research: Classification and considerations of use. Remote Sens. 2012, 4, 1671–1692. [Google Scholar] [CrossRef]
- Crebassol, P.; Ferrier, P.; Dedieu, G.; Hagolle, O.; Fougnie, B.; Tinto, F.; Yaniv, Y.; Herscovitz, J. VENµS (Vegetation and Environment Monitoring on a New Micro Satellite). In Small Satellite Missions for Earth Observation; Springer: Berlin, Germany, 2010; pp. 47–65. [Google Scholar]
- Laliberte, A.S.; Goforth, M.A.; Steele, C.M.; Rango, A. Multispectral remote sensing from unmanned aircraft: Image processing workflows and applications for rangeland environments. Remote Sens. 2011, 3, 2529–2551. [Google Scholar] [CrossRef]
- Blaschke, T.; Johansen, K.; Tiede, D. Object-Based Image Analysis for Vegetation Mapping and Monitoring. In Advances in Environmental Remote Sensing: Sensor, Algorithms, and Applications; CRC Press: Boca Raton, FL, USA, 2011; pp. 241–271. [Google Scholar]
- Haralick, R.M.; Shanmugam, K. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Schmidt, T.; Schuster, C.; Kleinschmit, B.; Forster, M. Evaluating an Intra-Annual Time Series for Grassland Classification—How Many Acquisitions and What Seasonal Origin Are Optimal? IEEE J. STARS 2014, 7, 3428–3439. [Google Scholar] [CrossRef]
- Schuster, C.; Schmidt, T.; Conrad, C.; Kleinschmit, B.; Förster, M. Grassland habitat mapping by intra-annual time series analysis–Comparison of RapidEye and TerraSAR-X satellite data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 25–34. [Google Scholar] [CrossRef]
- Lisein, J.; Michez, A.; Claessens, H.; Lejeune, P. Discrimination of deciduous tree species from time series of unmanned aerial system imagery. PLoS ONE 2015, 10, e0141006. [Google Scholar] [CrossRef] [PubMed]
- Michez, A.; Piégay, H.; Lisein, J.; Claessens, H.; Lejeune, P. Classification of riparian forest species and health condition using multi-temporal and hyperspatial imagery from unmanned aerial system. Environ. Monit. Assess. 2016, 188, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Cole, B.; McMorrow, J.; Evans, M. Spectral monitoring of moorland plant phenology to identify a temporal window for hyperspectral remote sensing of peatland. ISPRS J. Photogramm. Remote Sens. 2014, 90, 49–58. [Google Scholar] [CrossRef]
- Feilhauer, H.; Thonfeld, F.; Faude, U.; He, K.S.; Rocchini, D.; Schmidtlein, S. Assessing floristic composition with multispectral sensors—A comparison based on monotemporal and multiseasonal field spectra. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 218–229. [Google Scholar] [CrossRef]
- Féret, J.B.; Corbane, C.; Alleaume, S. Detecting the phenology and discriminating Mediterranean natural habitats with multispectral sensors—An analysis based on multiseasonal field spectra. IEEE J. STARS 2015, 8, 2294–2305. [Google Scholar] [CrossRef]
- Van Deventer, H.; Cho, M.A.; Mutanga, O. Improving the classification of six evergreen subtropical tree species with multi-season data from leaf spectra simulated to WorldView-2 and RapidEye. Int. J. Remote Sens. 2017, 38, 4804–4830. [Google Scholar] [CrossRef]
- Richardson, A.D.; Jenkins, J.P.; Braswell, B.H.; Hollinger, D.Y.; Ollinger, S.V.; Smith, M.L. Use of digital webcam images to track spring green-up in a deciduous broadleaf forest. Oecologia 2007, 152, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Sonnentag, O.; Hufkens, K.; Teshera-Sterne, C.; Young, A.M.; Friedl, M.; Braswell, B.H.; Richardson, A.D. Digital repeat photography for phenological research in forest ecosystems. Agric. For. Meteorol. 2012, 152, 159–177. [Google Scholar] [CrossRef]
- Wingate, L.; Ogée, J.; Cremonese, E.; Filippa, G.; Mizunuma, T.; Migliavacca, M.; Grace, J. Interpreting canopy development and physiology using a European phenology camera network at flux sites. Biogeosciences 2015, 12, 5995–6015. [Google Scholar] [CrossRef]
- Almeida, J.; dos Santos, J.A.; Alberton, B.; Morellato, L.P.C.; Torres, R.D.S. Phenological visual rhythms: Compact representations for fine-grained plant species identification. Pattern Recognit. Lett. 2016, 81, 90–100. [Google Scholar] [CrossRef]
- Ide, R.; Oguma, H. Use of digital cameras for phenological observations. Ecol. Inform. 2010, 5, 339–347. [Google Scholar] [CrossRef]
- Bater, C.W.; Coops, N.C.; Wulder, M.A.; Hilker, T.; Nielsen, S.E.; McDermid, G.; Stenhouse, G.B. Using digital time-lapse cameras to monitor species-specific understorey and overstorey phenology in support of wildlife habitat assessment. Environ. Monit. Assess. 2011, 180, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Snyder, K.A.; Wehan, B.L.; Filippa, G.; Huntington, J.L.; Stringham, T.K.; Snyder, D.K. Extracting Plant Phenology Metrics in a Great Basin Watershed: Methods and Considerations for Quantifying Phenophases in a Cold Desert. Sensors 2016, 16, 1948. [Google Scholar] [CrossRef] [PubMed]
- Danin, A. Flora and vegetation of Israel and adjacent areas. In The Zoogeography of Israel; Dr. W. Junk Publishers: Dordrecht, The Netherlands, 1988; pp. 251–276. [Google Scholar]
- Shmida, A. Mediterranean vegetation in California and Israel: Similarities and differences. Isr. J. Bot. 1981, 30, 105–123. [Google Scholar]
- Miller, P.C. Canopy structure of Mediterranean-type shrubs in relation to heat and moisture. In Mediterranean-Type Ecosystems; Springer: Berlin/Heidelberg, Germany, 1983; pp. 133–166. [Google Scholar]
- Ne’eman, G.; Goubitz, S. Phenology of east Mediterranean vegetation. In Life and Environment in the Mediterranean; WIT Press: Ashurst, UK, 2000; pp. 155–201. [Google Scholar]
- Orshan, G. Approaches to the definition of Mediterranean growth forms. In Mediterranean-Type Ecosystems; Springer: Berlin/Heidelberg, Germany, 1983; pp. 86–100. [Google Scholar]
- Kadmon, R.; Harari-Kremer, R. Studying long-term vegetation dynamics using digital processing of historical aerial photographs. Remote Sens. Environ. 1999, 68, 164–176. [Google Scholar] [CrossRef]
- Levin, N. Human factors explain the majority of MODIS-derived trends in vegetation cover in Israel: A densely populated country in the eastern Mediterranean. Reg. Environ. Chang. 2016, 16, 1197–1211. [Google Scholar] [CrossRef]
- Naveh, Z. Mediterranean landscape evolution and degradation as multivariate biofunctions: Theoretical and practical implications. Landsc. Plan 1982, 9, 125–146. [Google Scholar] [CrossRef]
- Perevolotsky, A.; Seligman, N.A.G. Role of grazing in Mediterranean rangeland ecosystems. Bioscience 1998, 48, 1007–1017. [Google Scholar] [CrossRef]
- Mandelik, Y.; Roll, U.; Fleischer, A. Cost-efficiency of biodiversity indicators for Mediterranean ecosystems and the effects of socio-economic factors. J. Appl. Ecol. 2010, 47, 1179–1188. [Google Scholar] [CrossRef]
- Radford, E.A.; Catullo, G.; de Montmollin, B. Important plant areas of the south and east Mediterranean region: Priority sites for conservation. International Union for Conservation of Nature, Plantlife, WWF, 2011. Available online: https://portals.iucn.org/library/sites/library/files/documents/2011-014.pdf (accessed on 17 September 2017).
- Manakos, I.; Manevski, K.; Petropoulos, G.P.; Elhag, M.; Kalaitzidis, C. Development of a spectral library for Mediterranean land cover types. In Proceedings of the 30th EARSeL Symposium: Remote Sensing for Science, Education and Natural and Cultural Heritage, Paris, France, 31 May–4 June 2010. [Google Scholar]
- Manevski, K.; Manakos, I.; Petropoulos, G.P.; Kalaitzidis, C. Discrimination of common Mediterranean plant species using field spectroradiometry. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 922–933. [Google Scholar] [CrossRef]
- Rud, R.; Shoshany, M.; Alchanatis, V.; Cohen, Y. Application of spectral features’ ratios for improving classification in partially calibrated hyperspectral imagery: A case study of separating Mediterranean vegetation species. J. Real Time Image Process. 2006, 1, 143–152. [Google Scholar] [CrossRef]
- Weil, G.; Lensky, I.M.; Levin, N. Using ground observations of a digital camera in the VIS-NIR range for quantifying the phenology of Mediterranean woody species. Int. J. Appl. Earth Obs. Geoinf. 2017, 62, 88–101. [Google Scholar] [CrossRef]
- Bar Massada, A.; Kent, R.; Blank, L.; Perevolotsky, A.; Hadar, L.; Carmel, Y. Automated segmentation of vegetation structure units in a Mediterranean landscape. Int. J. Remote Sens. 2012, 33, 346–364. [Google Scholar] [CrossRef]
- Bashan, D.; Bar-Massada, A. Regeneration dynamics of woody vegetation in a Mediterranean landscape under different disturbance-based management treatments. Appl. Veg. Sci. 2017, 20, 106–114. [Google Scholar] [CrossRef]
- Carmel, Y.; Kadmon, R. Computerized classification of Mediterranean vegetation using panchromatic aerial photographs. J. Veg. Sci. 1998, 9, 445–454. [Google Scholar] [CrossRef]
- Shoshany, M. Satellite remote sensing of natural Mediterranean vegetation: A review within an ecological context. Prog. Phys. Geogr. 2000, 24, 153–178. [Google Scholar] [CrossRef]
- Džubáková, K.; Molnar, P.; Schindler, K.; Trizna, M. Monitoring of riparian vegetation response to flood disturbances using terrestrial photography. Hydrol. Earth Syst. Sci. 2015, 19, 195–208. [Google Scholar] [CrossRef]
- Rabatel, G.; Gorretta, N.; Labbe, S. Getting simultaneous red and near-infrared band data from a single digital camera for plant monitoring applications: Theoretical and practical study. Biosyst. Eng. 2014, 117, 2–14. [Google Scholar] [CrossRef]
- Yang, C.; Westbrook, J.K.; Suh, C.P.C.; Martin, D.E.; Hoffmann, W.C.; Lan, Y.; Goolsby, J.A. An airborne multispectral imaging system based on two consumer-grade cameras for agricultural remote sensing. Remote Sens. 2014, 6, 5257–5278. [Google Scholar] [CrossRef]
- Nevo, E.; Fragman, O.; Dafni, A.; Beiles, A. Biodiversity and interslope divergence of vascular plants caused by microclimatic differences at “Evolution Canyon”, Lower Nahal Oren, Mount Carmel, Israel. Isr. J. Plant Sci. 1999, 47, 49–59. [Google Scholar] [CrossRef]
- Nijland, W.; de Jong, R.; de Jong, S.M.; Wulder, M.A.; Bater, C.W.; Coops, N.C. Monitoring plant condition and phenology using infrared sensitive consumer grade digital cameras. Agric. For. Meteorol. 2014, 184, 98–106. [Google Scholar] [CrossRef]
- Osaki, K. Appropriate luminance for estimating vegetation index from digital camera images. Bull. Soc. Sci. Photogr. Jpn. 2015, 25, 31–37. [Google Scholar]
- Hadjimitsis, D.G.; Clayton, C.R.I.; Retalis, A. The use of selected pseudo-invariant targets for the application of atmospheric correction in multi-temporal studies using satellite remotely sensed imagery. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 192–200. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASABE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Richardson, A.D.; Braswell, B.H.; Hollinger, D.Y.; Jenkins, J.P.; Ollinger, S.V. Near-surface remote sensing of spatial and temporal variation in canopy phenology. Ecol. Appl. 2009, 19, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tang, J.; Mustard, J.F. Beyond leaf color: Comparing camera-based phenological metrics with leaf biochemical, biophysical, and spectral properties throughout the growing season of a temperate deciduous forest. J. Geophys. Res. Biogeosci. 2014, 119, 181–191. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Peña, J.M.; De Castro, A.I.; López-Granados, F. Multi-temporal mapping of the vegetation fraction in early-season wheat fields using images from UAV. Comput. Electron. Agric. 2014, 103, 104–113. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Soudani, K.; Hmimina, G.; Delpierre, N.; Pontailler, J.Y.; Aubinet, M.; Bonal, D.; Caquetd, B.; de Grandcourtd, A.; Burbane, B.; Flechard, C.; et al. Ground-based Network of NDVI measurements for tracking temporal dynamics of canopy structure and vegetation phenology in different biomes. Remote Sens. Environ. 2012, 123, 234–245. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Lebourgeois, V.; Bégué, A.; Labbé, S.; Mallavan, B.; Prévot, L.; Roux, B. Can commercial digital cameras be used as multispectral sensors? A crop monitoring test. Sensors 2008, 8, 7300–7322. [Google Scholar] [CrossRef] [PubMed]
- Motohka, T.; Nasahara, K.N.; Oguma, H.; Tsuchida, S. Applicability of green-red vegetation index for remote sensing of vegetation phenology. Remote Sens. 2010, 2, 2369–2387. [Google Scholar] [CrossRef]
- Nagai, S.; Saitoh, T.M.; Kobayashi, H.; Ishihara, M.; Suzuki, R.; Motohka, T.; Nasahara, K.N.; Muraoka, H. In situ examination of the relationship between various vegetation indices and canopy phenology in an evergreen coniferous forest, Japan. Int. J. Remote Sens. 2012, 33, 6202–6214. [Google Scholar] [CrossRef]
- Almeida, J.; dos Santos, J.A.; Alberton, B.; Torres, R.D.S.; Morellato, L.P.C. Remote phenology: Applying machine learning to detect phenological patterns in a cerrado savanna. In Proceedings of the IEEE 8th International Conference, Chicago, IL, USA, 8–12 October 2012. [Google Scholar]
- Joblove, G.H.; Greenberg, D. Colour spaces for computer graphics. In Proceedings of the 5th Annual Conference on Computer Graphics and Interactive Techniques, Atlanta, GA, USA, 23–25 August 1978. [Google Scholar]
- Crimmins, M.A.; Crimmins, T.M. Monitoring plant phenology using digital repeat photography. J. Environ. Manag. 2008, 41, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Graham, E.A.; Yuen, E.M.; Robertson, G.F.; Kaiser, W.J.; Hamilton, M.P.; Rundel, P.W. Budburst and leaf area expansion measured with a novel mobile camera system and simple color thresholding. Environ. Exp. Bot. 2009, 65, 238–244. [Google Scholar] [CrossRef]
- Mizunuma, T.; Mencuccini, M.; Wingate, L.; Ogée, J.; Nichol, C.; Grace, J. Sensitivity of colour indices for discriminating leaf colours from digital photographs. Methods Ecol. Evol. 2014, 5, 1078–1085. [Google Scholar] [CrossRef]
- Cleveland, W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Gómez, C.; White, J.C.; Wulder, M.A. Optical remotely sensed time series data for land cover classification: A review. ISPRS J. Photogramm. Remote Sens. 2016, 116, 55–72. [Google Scholar] [CrossRef]
- Löw, F.; Conrad, C.; Michel, U. Decision fusion and non-parametric classifiers for land use mapping using multi-temporal RapidEye data. ISPRS J. Photogramm. Remote Sens. 2015, 108, 191–204. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Sheffer, E.; Canham, C.D.; Kigel, J.; Perevolotsky, A. An integrative analysis of the dynamics of landscape-and local-scale colonization of Mediterranean woodlands by Pinus halepensis. PLoS ONE 2014, 9, e90178. [Google Scholar] [CrossRef] [PubMed]
- Tessler, N.; Wittenberg, L.; Provizor, E.; Greenbaum, N. The influence of short-interval recurrent forest fires on the abundance of Aleppo pine (Pinus halepensis Mill.) on Mount Carmel, Israel. For. Ecol. Manag. 2014, 324, 109–116. [Google Scholar] [CrossRef]
- Ahmed, O.S.; Shemrock, A.; Chabot, D.; Dillon, C.; Williams, G.; Wasson, R.; Franklin, S.E. Hierarchical land cover and vegetation classification using multispectral data acquired from an unmanned aerial vehicle. Int. J. Remote Sens. 2017, 38, 1–16. [Google Scholar]
- Von Bueren, S.K.; Burkart, A.; Hueni, A.; Rascher, U.; Tuohy, M.P.; Yule, I.J. Deploying four optical UAV-based sensors over grassland: Challenges and limitations. Biogeosciences 2015, 12, 163. [Google Scholar] [CrossRef]
- Darwish, A.; Leukert, K.; Reinhardt, W. Image segmentation for the purpose of object-based classification. In Proceedings of the International Geoscience and Remote Sensing Symposium, Toulouse, France, 21–25 July 2003. [Google Scholar]
- Hájek, F. Object-oriented classification of remote sensing data for the identification of tree species composition. In Proceedings of the ForestSat Conference, Borås, Sweden, 31 May–3 June 2005. [Google Scholar]
- Kaszta, Ż.; Van De Kerchove, R.; Ramoelo, A.; Cho, M.A.; Madonsela, S.; Mathieu, R.; Wolff, E. Seasonal Separation of African Savanna Components Using Worldview-2 Imagery: A Comparison of Pixel-and Object-Based Approaches and Selected Classification Algorithms. Remote Sens. 2016, 8, 763. [Google Scholar] [CrossRef]
- Lehmann, J.R.; Münchberger, W.; Knoth, C.; Blodau, C.; Nieberding, F.; Prinz, T.; Kleinebecker, T. High-Resolution Classification of South Patagonian Peat Bog Microforms Reveals Potential Gaps in Up-Scaled CH4 Fluxes by use of Unmanned Aerial System (UAS) and CIR Imagery. Remote Sens. 2016, 8, 173. [Google Scholar] [CrossRef]
- Mallinis, G.; Koutsias, N.; Tsakiri-Strati, M.; Karteris, M. Object-based classification using Quickbird imagery for delineating forest vegetation polygons in a Mediterranean test site. ISPRS J. Photogramm. Remote Sens. 2008, 63, 237–250. [Google Scholar] [CrossRef]
- Baatz, M.; Schäpe, A. Multiresolution segmentation: An optimization approach for high quality multi-scale image segmentation. Angewandte Geographische Informationsverarbeitung 2000, 58, 12–23. [Google Scholar]
- Flanders, D.; Hall-Beyer, M.; Pereverzoff, J. Preliminary evaluation of eCognition object-based software for cut block delineation and feature extraction. Can. J Remote Sens. 2003, 29, 441–452. [Google Scholar] [CrossRef]
- Eastman, J.R.; Filk, M. Long sequence time series evaluation using standardized principal components. Photogramm. Eng. Remote Sens. 1993, 59, 991–996. [Google Scholar]
- Kuzmin, A.; Korhonen, L.; Manninen, T.; Maltamo, M. Automatic Segment-Level Tree Species Recognition Using High Resolution Aerial Winter Imagery. Eur. J Remote Sens. 2016, 49, 239–259. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Mafanya, M.; Tsele, P.; Botai, J.; Manyama, P.; Swart, B.; Monate, T. Evaluating pixel and object based image classification techniques for mapping plant invasions from UAV derived aerial imagery: Harrisia pomanensis as a case study. ISPRS J. Photogramm. Remote Sens. 2017, 129, 1–11. [Google Scholar] [CrossRef]
- Orshan, G. Plant Pheno-Morphological Studies in Mediterranean Type Ecosystems; Kluwer: Dordrecht, The Netherlands, 1989. [Google Scholar]
- Helman, D.; Lensky, I.M.; Tessler, N.; Osem, Y.A. Phenology-Based Method for Monitoring Woody and Herbaceous Vegetation in Mediterranean Forests from NDVI Time Series. Remote Sens. 2015, 7, 12314–12335. [Google Scholar] [CrossRef]
- Donnelly, A.; Yu, R.; Caffarra, A.; Hanes, J.; Liang, L.; Desai, A.R.; Desaie, A.R.; Liuf, L.; Schwartz, M.D. Interspecific and interannual variation in the duration of spring phenophases in a northern mixed forest. Agric. For. Meteorol. 2017, 243, 55–67. [Google Scholar] [CrossRef]
- Pinto, C.A.; Henriques, M.O.; Figueiredo, J.P.; David, J.S.; Abreu, F.G.; Pereira, J.S.; David, T.S. Phenology and growth dynamics in Mediterranean evergreen oaks: Effects of environmental conditions and water relations. For. Ecol. Manag. 2011, 262, 500–508. [Google Scholar] [CrossRef]
- Vitasse, Y.; Delzon, S.; Dufrêne, E.; Pontailler, J.Y.; Louvet, J.M.; Kremer, A.; Michalet, R. Leaf phenology sensitivity to temperature in European trees: Do within-species populations exhibit similar responses? Agric. For. Meteorol. 2009, 149, 735–744. [Google Scholar] [CrossRef]
- Fisher, J.I.; Mustard, J.F.; Vadeboncoeur, M.A. Green leaf phenology at Landsat resolution: Scaling from the field to the satellite. Remote Sens. Environ. 2006, 100, 265–279. [Google Scholar] [CrossRef]
- Gates, D.M.; Keegan, H.J.; Schleter, J.C.; Weidner, V.R. Spectral properties of plants. Appl. Opt. 1965, 4, 11–20. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Hufkens, K.; Friedl, M.; Sonnentag, O.; Braswell, B.H.; Milliman, T.; Richardson, A.D. Linking near-surface and satellite remote sensing measurements of deciduous broadleaf forest phenology. Remote Sens. Environ. 2012, 117, 307–321. [Google Scholar] [CrossRef]
- Müllerová, J.; Brůna, J.; Bartaloš, T.; Dvořák, P.; Vítková, M.; Pyšek, P. Timing Is Important: Unmanned Aircraft vs. Satellite Imagery in Plant Invasion Monitoring. Front. Plant Sci. 2017, 8, 887. [Google Scholar] [CrossRef] [PubMed]
- Butler, D. Many eyes on Earth. Nature 2014, 505, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Houborg, R.; McCabe, M.F. High-Resolution NDVI from Planet’s Constellation of Earth Observing Nano-Satellites: A New Data Source for Precision Agriculture. Remote Sens. 2016, 8, 768. [Google Scholar] [CrossRef]
- Sandau, R.; Brieß, K.; D’Errico, M. Small satellites for global coverage: Potential and limits. ISPRS J. Photogramm. Remote Sens. 2010, 65, 492–504. [Google Scholar] [CrossRef]
- Strauss, M. Planet Earth to get a daily selfie. Science 2017, 355, 782–783. [Google Scholar] [CrossRef] [PubMed]
- Dedieu, G.; Karnieli, A.; Hagolle, O.; Jeanjean, H.; Cabot, F.; Ferrier, P.; Yaniv, Y. VENµS: A joint Israel–French earth observation, scientific mission with high spatial and temporal resolution capabilities. In Proceedings of the Recent Advances in Quantitative Remote Sensing, Valencia, Spain, 25–29 September 2006. [Google Scholar]









| Modified Canon EOS 600D®, Near-Surface Observations | Micasense Rededge®, Overhead Observations | |
|---|---|---|
| Sensor | CMOS | Separate sensor for each band. Down-welling light sensor. |
| Bands | Blue, green, red and near-infrared. Visible bands and near-infrared band were produced with separate external filters. | Blue, green, red, red-edge and near-infrared. |
| Band width | Relatively wide and overlapping, see technical description of LCC-LDP© labs X-Nite CC1® and X-Nite 780® filters (https://www.maxmax.com/filters). | Relatively narrow and separate. 20 nm for blue and green, 10 nm for red and red-edge, 40 nm for near-infrared. |
| Pixel resolution | 18.7 Megapixel | 1.2 Megapixel. 8 cm per pixel at 120 m above ground level. |
| Radiometric resolution | 14-bit | 12-bit |
| Spectral Index | Formula | Reference | Explanation/Objective |
|---|---|---|---|
| Relative green/Green chromatic coordinate | [40,41,73,74,75] | The relative component of green, red and blue bands over the total sum of all camera bands. Less affected from scene illumination conditions than the original band values. | |
| Relative red/Red chromatic coordinate | |||
| Relative blue/Blue chromatic coordinate | |||
| Green excess/Excess green; ExG | [40,44,70] | Effective for distinction between green vegetation and soil. | |
| Green excess-Red excess; ExGR | [76,77] | Improvement of ExG, better distinction between vegetation and soil. | |
| Normalized Difference Vegetation Index; NDVI | [66,78,79] | Relationship between the NIR and red bands indicates vegetation condition due to chlorophyll absorption within red spectral range and high reflectance within the NIR range. | |
| Green Normalized Difference Vegetation Index; gNDVI | [80,81] | Improvement of NDVI, accurate in assessing chlorophyll content. | |
| Green-Red Vegetation Index; GRVI | [82,83] | Relationship between the green and red bands is an effective index for detecting phenophases. | |
| Total brightness | [35,40,84] | Can describe prominent visual changes in foliage (e.g., white flowering of Prunus dulcis). | |
| RGB conversion to Hue, Saturation and Value; HSV | See [85] | [85,86,87,88] | Alternative colour space for describing canopy changes. Compared to RGB-derived indices, can be more effective and robust as a proxy for leaf development. |
| Random Forest Optimal Dates (Ground-Based Preliminary Feature Selection Analysis, a Single Run of the Feature Selection Process) | Actual Overhead Data Acquisition Dates |
|---|---|
| 16 December 2015 | 20 December 2016 |
| 13 January 2016 | 16 January 2017 |
| 24 February 2016 | 25 February 2017 |
| 5 April 2016 | 10 April 2017 |
| 14 June 2016 | 18 June 2017 |
| Scenario | Color in Figure 9 | Explanation | Relative Red, Green and Blue | Hue, Saturation, Value | ExG, GRVI, EmE, Total Brightness | NDVI, gNDVI |
|---|---|---|---|---|---|---|
| 1 | ▬ | All spectral indices | V | V | V | V |
| 2 | ▬ | Spectral indices without NIR band | V | V | V | |
| 3 | ▬ | ExG * | ExG only | |||
| 4 | ▬ | HSV conversion only | V | |||
| 5 | ▬ | Relative red, green and blue only | V | |||
| 6 | ▬ | Spectral indices without NIR band and without HSV conversion | V | V |
| Classified | Reference | |||||||||||
| Winter Deciduous Broad-Leaved Tree | Evergreen Broad-Leaved Tree | Evergreen/Summer Semi-Deciduous Broad-Leaved Shrub | Green During Wet Period—Winter and Spring | Evergreen/Summer Semi-Deciduous Broad-Leaved Shrub | Evergreen Broad-Leaved Shrub | Evergreen Broad-Leaved Shrub | Evergreen Broad-Leaved Shrub | Evergreen/Summer Semi-Deciduous Broad-Leaved Shrub | ||||
| Prunus dulcis | Quercus calliprinos | Rhamnus lycioides | Herbaceous patches | Cistus creticus/salviifolius | Pistacia lentiscus | Olea europaea | Pinus halepensis | Sarcopoterium spinosum | Total Individuals 1 | Average User’s Accuracy | ||
| Prunus dulcis | 86.3% | 0% | 10.0% | 0.4% | 0% | 0% | 0% | 0% | 0% | 57 | 88.5 | |
| Quercus calliprinos | 1.8% | 82.2% | 0% | 0.7% | 3.4% | 9.8% | 0% | 6.2% | 0% | 260 | 87.5 | |
| Rhamnus lycioides | 11.2% | 0.4% | 83.4% | 0.1% | 0% | 1.1% | 5.3% | 0% | 0.6% | 58 | 78.6 | |
| Herbaceous patches | 0% | 0% | 0% | 94.2% | 0% | 0% | 0% | 0% | 6.2% | 151 | 97.1 | |
| Cistus creticus/salviifolius | 0% | 0.2% | 0.3% | 1.1% | 85.4% | 1.4% | 0.4% | 0.8% | 5.3% | 41 | 78.1 | |
| Pistacia lentiscus | 0% | 13.7% | 1.0% | 0% | 4.9% | 81.6% | 17.3% | 2.3% | 0% | 244 | 80.8 | |
| Olea europaea | 0.4% | 0.5% | 4.8% | 0% | 1.5% | 5.7% | 76.9% | 0% | 0% | 45 | 64.6 | |
| Pinus halepensis | 0% | 3.1% | 0% | 0% | 2.4% | 0.2% | 0% | 90.8% | 0% | 51 | 83.1 | |
| Sarcopoterium spinosum | 0.4% | 0% | 0.3% | 3.6% | 2.4% | 0% | 0% | 0% | 87.9% | 68 | 89.8 | |
| Total individuals 1 | 57 | 260 | 58 | 151 | 41 | 244 | 45 | 51 | 68 | 972 | ||
| Average producer’s Accuracy | 86.3 | 82.2 | 83.5 | 94.2 | 85.4 | 81.6 | 76.9 | 90.8 | 87.9 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weil, G.; Lensky, I.M.; Resheff, Y.S.; Levin, N. Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection. Remote Sens. 2017, 9, 1130. https://doi.org/10.3390/rs9111130
Weil G, Lensky IM, Resheff YS, Levin N. Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection. Remote Sensing. 2017; 9(11):1130. https://doi.org/10.3390/rs9111130
Chicago/Turabian StyleWeil, Gilad, Itamar M. Lensky, Yehezkel S. Resheff, and Noam Levin. 2017. "Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection" Remote Sensing 9, no. 11: 1130. https://doi.org/10.3390/rs9111130
APA StyleWeil, G., Lensky, I. M., Resheff, Y. S., & Levin, N. (2017). Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection. Remote Sensing, 9(11), 1130. https://doi.org/10.3390/rs9111130








