An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index
Abstract
:1. Introduction
2. Material and Methods
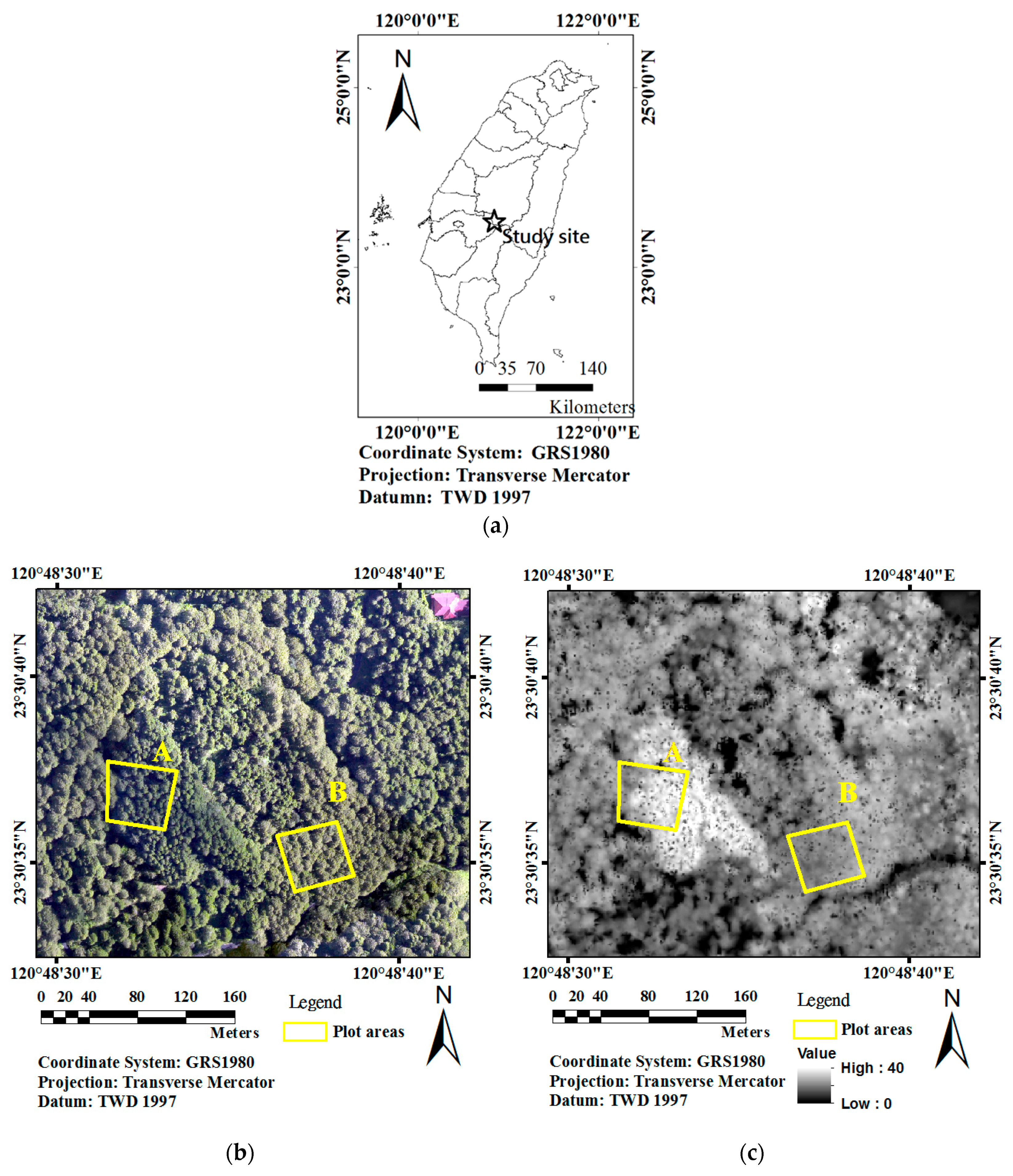
2.1. Airborne LiDAR Data Collection
2.2. Datasets Used to Train and Validate the AGC Models
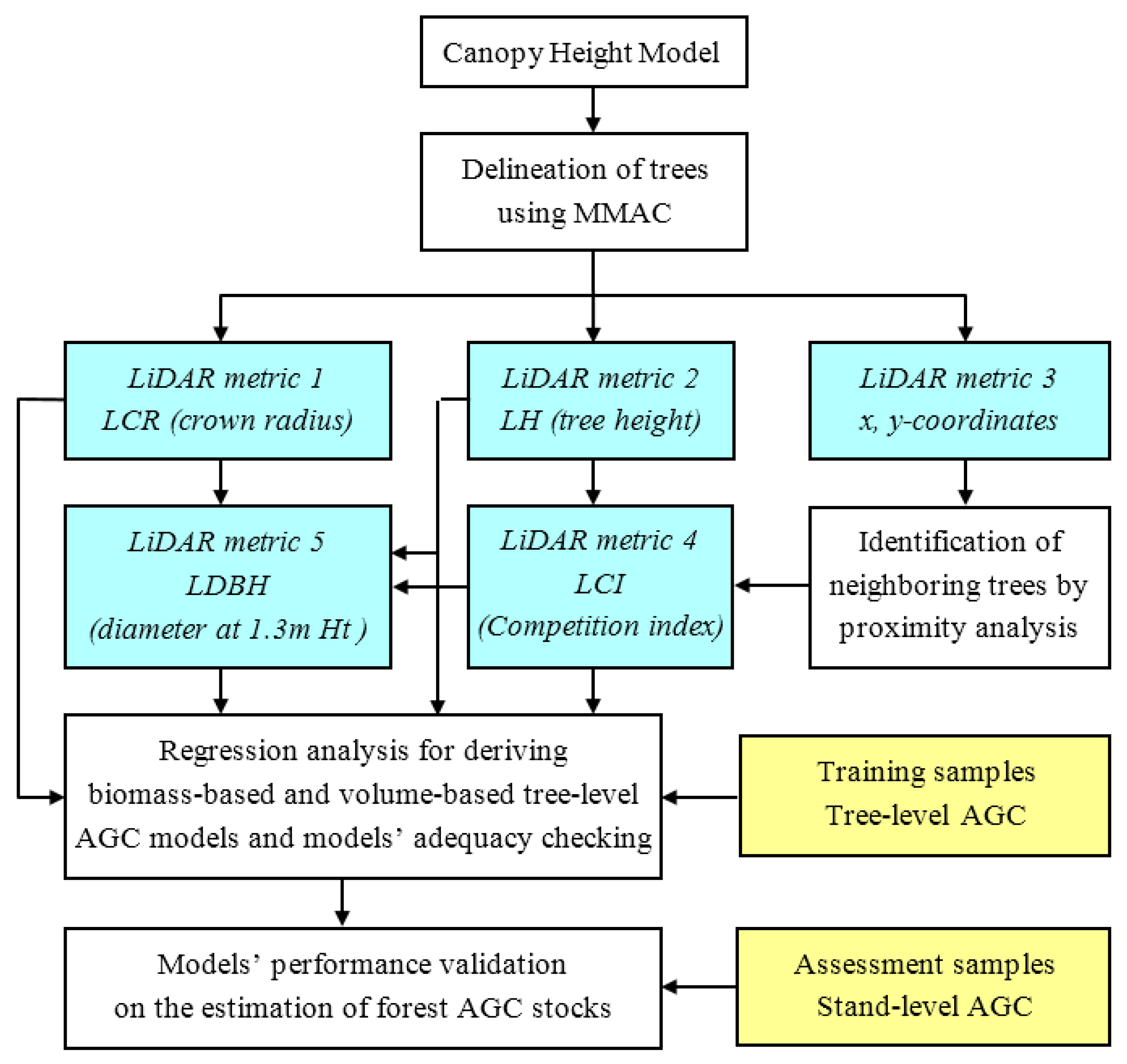
2.3. A Tree-Level Algorithm to Account for Above-Ground Carbon
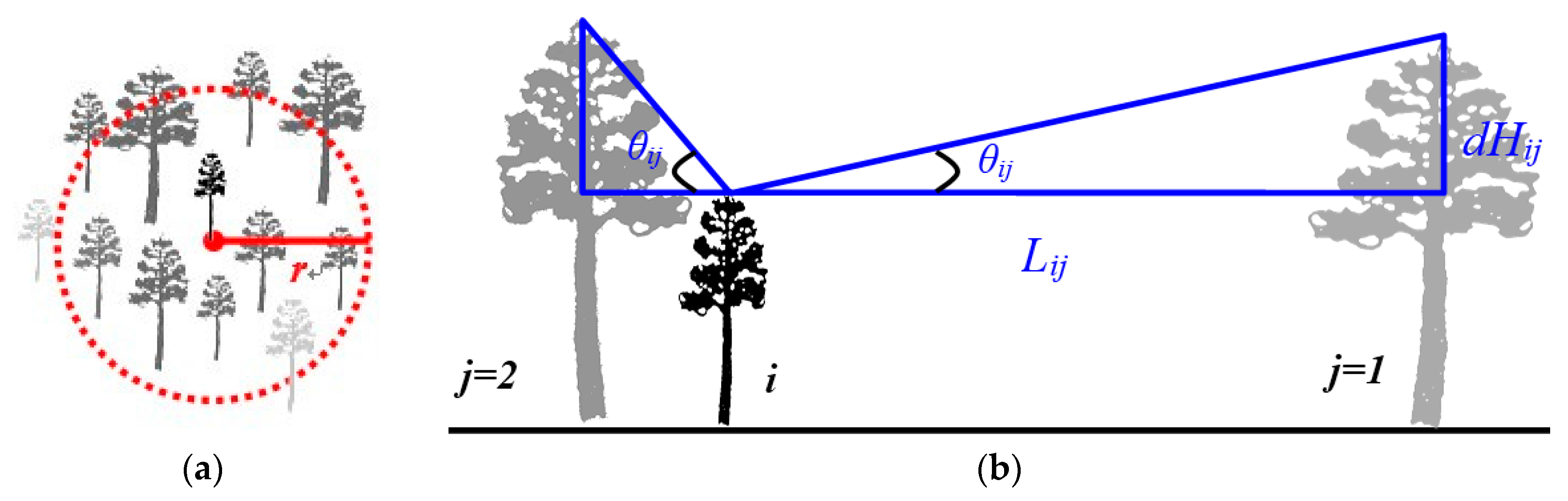
2.3.1. LiDAR Derived Tree-Level Parameters
2.3.2. Regression Analysis for Deriving Tree-Level AGC Models
2.3.3. Evaluating the Predictive Performance of the Models
3. Results
3.1. LiDAR-Based Tree-Level AGC Models
3.2. Performance Comparison of the LiDAR-Based Tree-Level AGC Models
4. Discussion
4.1. A Post-Hoc Examination of the Prediction Accuracy of AGC Stock Estimation at Stand-Level
4.2. Detrimental Combinations of LiDAR-Derived Tree Parameters and Increased Uncertainty in the Estimation of Stand-Level AGC
4.3. Why Use Biomass-Based or Volume-Based AGC Models in Predicting Forest Carbon Stock?
- (1)
- biomass-based model C2: AGC = 0.000068159 × LDBH1.4299 × LH1.1708 × LCI−0.0573,
- (2)
- volume-based model V4M: AGC = [0.2919(a × LDBHb × LHc)1.0026], and
- (3)
- volume-based model V5: AGC = [exp(−6.2803 + 2.3774lnLH − 0.0145LCI + 0.0316LCR)] × D × BEF × CF.
4.4. Recommendations for Future Work
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lin, C.; Lin, C.H. Comparison of carbon sequestration potential in agricultural and afforestation farming systems. Sci. Agricola 2013, 70, 93–101. [Google Scholar] [CrossRef]
- Tsogt, K.; Lin, C. A flexible modeling of irregular diameter structure for the volume estimation of forest stands. J. For. Res. 2014, 19, 1–11. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Scrivani, J.A. Fusion of small-footprint LiDAR and multispectral data to estimate plot-level volume and biomass in deciduous and pine forests in Virginia, USA. For. Sci. 2004, 50, 551–565. [Google Scholar]
- Almeida, A.C.; Barros, P.L.C.; Monteiro, J.H.A.; Rocha, B.R.P. Estimation of above-ground forest biomass in Amazonia with neural networks and remote sensing. IEEE Lat. Am. Trans. 2009, 7, 27–32. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S.; Nelson, R. LiDAR remote sensing of forest biomass: A scale-invariant estimation approach using airborne lasers. Remote Sens. Environ. 2009, 113, 182–196. [Google Scholar] [CrossRef]
- Badreldin, N.; Sanchez-Azofeifa, A. Estimating forest biomass dynamics by integrating multi-temporal Landsat satellite images with ground and airborne LiDAR data in the Coal Valley Mine, Alberta, Canada. Remote Sens. 2015, 7, 2832–2849. [Google Scholar] [CrossRef]
- Hansen, E.H.; Gobakken, T.; Bollandsås, O.M.; Zahabu, E.; Næsset, E. Modeling aboveground biomass in dense tropical submontane rainforest using airborne laser scanner data. Remote Sens. 2015, 7, 788–807. [Google Scholar] [CrossRef]
- Sheridan, R.D.; Popescu, S.C.; Gatziolis, D.; Morgan, C.L.S.; Ku, N.W. Modeling forest aboveground biomass and volume using airborne LiDAR metrics and forest inventory and analysis data in the Pacific Northwest. Remote Sens. 2015, 7, 229–255. [Google Scholar] [CrossRef]
- Meyer, V.; Saatchi, S.S.; Chave, J.; Dalling, J.W.; Bohlman, S.; Fricker, G.A.; Robinson, C.; Neumann, M.; Hubbell, S. Detecting tropical forest biomass dynamics from repeated airborne LiDAR measurements. Biogeosciences 2013, 10, 5421–5438. [Google Scholar] [CrossRef]
- Simonson, W.; Ruiz-Benito, P.; Valladares, F.; Coomes, D. Modelling above-ground carbon dynamics using multi-temporal airborne LiDAR: Insights from a Mediterranean woodland. Biogeosciences 2016, 13, 961–973. [Google Scholar] [CrossRef]
- Jubanski, J.; Ballhorn, U.; Kronseder, K.; Franke, J.; Siegert, F. Detection of large above-ground biomass variability in lowland forest ecosystems by airborne LiDAR. Biogeosciences 2013, 10, 3917–3930. [Google Scholar] [CrossRef]
- Lefsky, M.A. A global forest canopy height map from the Moderate Resolution Imaging Spectroradiometer and the Geoscience Laser Altimeter System. Geophys. Res. Lett. 2010, 37, L15401. [Google Scholar] [CrossRef]
- Popescu, S.C.; Zhao, K.; Neuenschwander, A.; Lin, C. Satellite LiDAR vs. small footprint airborne LiDAR: Comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens. Environ. 2011, 115, 2786–2797. [Google Scholar] [CrossRef]
- Los, S.O.; Rosette, J.A.B.; Kljun, N.; North, P.R.J.; Chasmer, L.; Suarez, J.C.; Hopkinson, C.; Hill, R.A.; van Gorsel, E.; Mahoney, C.; et al. Vegetation height and cover fraction between 60°S and 60°N from ICESat GLAS data. Geosci. Model Dev. 2012, 5, 413–432. [Google Scholar] [CrossRef]
- Baccini, A.; Goetz, S.J.; Walker, W.S.; Laporte, N.T.; Sun, M.; Sulla-Menashe, D.; Hackler, J.; Beck, P.S.A.; Dubayah, R.; Friedl, M.A.; et al. Estimated carbon dioxide emissions from tropical deforestation improved by carbon density maps. Nat. Clim. Chang. 2012, 2, 182–185. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.A.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Gaveau, L.A.; Hill, R.A. Quantifying canopy height underestimation by laser pulse penetration in small-footprint airborne laser scanning data. Can. J. Remote Sens. 2003, 29, 650–657. [Google Scholar] [CrossRef]
- Magnussen, S.; Nasset, E.; Gobakken, T. Reliability of LiDAR derived predictors of forest inventory attributes: A case study with Norway spruce. Remote Sens. Environ. 2010, 114, 700–712. [Google Scholar] [CrossRef]
- Niska, H.; Skon, J.P.; Packalen, P.; Tokola, T.; Maltamo, M.; Kolehmainen, M. Neural networks for the prediction of species-specific plot volumes using airborne laser scanning and aerial photographs. IEEE Trans. Geosci. Remote Sens. 2010, 48, 1076–1085. [Google Scholar] [CrossRef]
- Tesfamichael, S.G.; van Aardt, J.A.N.; Ahmed, F. Estimating plot-level tree height and volume of Eucalyptus grandis plantations using small-footprint, discrete return LiDAR data. Progress Phys. Geogr. 2010, 34, 515–540. [Google Scholar] [CrossRef]
- Clark, D.B.; Clark, D.A. Landscape-scale variation in forest structure and biomass in a tropical rain forest. For. Ecol. Manag. 2000, 137, 185–198. [Google Scholar] [CrossRef]
- Bombelli, A.; Avitabile, V.; Balzter, H.; Marchesini, L.B.; Bernoux, M.; Brady, M.; Hall, R.; Hansen, M.; Henry, M.; Herold, M.; et al. Biomass—Assessment of the Status of the Development of the Standards for the Terrestrial Essential Climate Variables; Global Terrestrial Observing System, Food and Agricultural Organization of United Nations: Rome, Italy, 2009. [Google Scholar]
- Tsui, O.W.; Coops, N.C.; Wulder, M.A.; Marshall, P.L.; McCardle, A. Using multi-frequency radar and discrete-return LiDAR measurements to estimate above-ground biomass and biomass components in a coastal temperate forest. ISPRS J. Photogramm. Remote Sens. 2012, 69, 121–133. [Google Scholar] [CrossRef]
- Maselli, F.; Chiesi, M.; Montaghi, A.; Pranzini, E. Use of ETM+ images to extend stem volume estimates obtained from LiDAR data. ISPRS J. Photogramm. Remote Sens. 2011, 66, 662–671. [Google Scholar] [CrossRef]
- Zhao, F.; Guo, Q.; Kellya, M. Allometric equation choice impacts LiDAR-based forest biomass estimates: A case study from the Sierra National Forest, CA. Agric. For. Meteorol. 2012, 165, 64–72. [Google Scholar] [CrossRef]
- Jakubowski, M.K.; Li, W.; Guo, Q.; Kelly, M. Delineating individual trees from LiDAR data: A comparison of vector- and raster-based segmentation approaches. Remote Sens. 2013, 5, 4163–4186. [Google Scholar] [CrossRef]
- Popescu, S.C. Estimating biomass of individual pine trees using airborne LiDAR. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]
- Chen, Q.; Gong, P.; Baldocchi, D.; Tian, Y.Q. Estimating basal area and stem volume for individual trees from LiDAR data. Photogramm. Eng. Remote Sens. 2007, 73, 1355–1365. [Google Scholar] [CrossRef]
- Dean, T.J.; Cao, Q.V.; Roberts, S.D.; Evans, D.L. Measuring heights to crown base and crown median with LiDAR in a mature, even-aged loblolly pine stand. For. Ecol. Manag. 2009, 257, 126–133. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. A system for the estimation of single-tree stem diameter and volume using multireturn LiDAR data. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2479–2490. [Google Scholar] [CrossRef]
- Dalponte, M.; Coops, N.C.; Bruzzone, L.; Gianelle, D. Aanlysis on the use of multiple returns LiDAR data for the estimation of tree stems volume. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2009, 2, 310–318. [Google Scholar] [CrossRef]
- Barilotti, A.; Sepic, F. Assessment of forestry parameters at single-tree level by using methods of LiDAR data analysis and processing. Ambiência 2010, 6, 81–92. [Google Scholar]
- Lin, C.; Lo, C.S.; Thomson, G. Estimating individual tree characteristics using the MMAC algorithm and a LiDAR -derived canopy height model. J. Earth Sci. Eng. 2011, 1, 35–41. [Google Scholar]
- Lo, C.S.; Lin, C. Growth-competition-based stem diameter and volume modeling for tree-level forest inventory using airborne LiDAR Data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2216–2226. [Google Scholar] [CrossRef]
- Forest Resources Assessment FAO. Global Forest Resources Assessment 2015—How Are the World’s Forests Changing; Food and Agricultural Organization of United Nations: Rome, Italy, 2015. [Google Scholar]
- IPCC. Good Practice Guidance for Land Use, Land-Use Change and Forestry; IPCC/OECD/IEA/IGES: Hayama, Japan, 2003. [Google Scholar]
- Lin, C.; Thomson, G.; Lo, C.S.; Yang, M.S. A multi-level morphological active contour algorithm for delineating tree crowns in mountainous forest. Photogramm. Eng. Remote Sens. 2011, 77, 241–249. [Google Scholar] [CrossRef]
- Wang, C.H.; Feng, F.L.; Lin, C.; Wang, Y.C.; Wang, Y.N.; Lin, S.T.; Chiou, C.R.; Yen, C.H.; Chung, Y.L.; Liu, C.P.; et al. Constructing Models for Transforming Forest Stock into Biomass (1/3); Technical Report No. 95AS-12.3.5-e2; Taiwan Forestry Bureau: Taipei, Taiwan, 2006.
- Hyndman, R.; Koehler, A. Another look at measures of forecast accuracy. Int. J. For. 2006, 22, 679–688. [Google Scholar] [CrossRef]
- Lin, C.; Dugarsuren, N. Deriving the spatiotemporal NPP pattern in terrestrial ecosystems of Mongolia using MODIS imagery. Photogramm. Eng. Remote Sens. 2015, 81, 587–598. [Google Scholar] [CrossRef]
- Duncan, D.B. Multiple range and multiple F tests. Biometrics 1955, 11, 1–42. [Google Scholar] [CrossRef]
- Montgomery, D.C. Design and Analysis of Experiments, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1984; pp. 66–68. [Google Scholar]
- Tomppo, E.; Gschwantner, T.; Lawrence, M.; McRoberts, R.E. National Forest Inventories—Pathways for Common Reporting; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Santoro, M.; Eriksson, L.; Fransson, J. Reviewing ALOS PALSAR backscatter observations for stem volume retrieval in Swedish forest. Remote Sens. 2015, 7, 4290–4317. [Google Scholar] [CrossRef]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Hosoi, F.; Nakai, Y.; Omasa, K. 3-D voxel-based solid modeling of a broad-leaved tree for accurate volume estimation using portable scanning LiDAR. ISPRS J. Photogramm. Remote Sens. 2013, 82, 41–48. [Google Scholar] [CrossRef]
- Kankare, V.; Holopainen, M.; Vastaranta, M.; Puttonen, E.; Yu, X.; Hyyppä, J.; Vaaja, M.; Hyyppä, H.; Alho, P. Individual tree biomass estimation using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2013, 75, 64–75. [Google Scholar] [CrossRef]
- Abdullahi, S.; Kugler, F.; Pretzsch, H. Prediction of stem volume in complex temperate forest stands using TanDEM-X SAR data. Remote Sens. Environ. 2016, 174, 197–211. [Google Scholar] [CrossRef]
- Zolkos, S.G.; Goetz, S.J.; Dubayah, R. A meta-analysis of terrestrial aboveground biomass estimation using LiDAR remote sensing. Remote Sens. Environ. 2013, 281, 289–298. [Google Scholar] [CrossRef]
- Marklund, L.G.; Schoene, D. Global assessment of growing stock, biomass and carbon stock. In Forest Resources Assessment Programme Working Paper 106/E; FAO: Rome, Italy, 2006; p. 55. [Google Scholar]
- IPCC. Guidelines for National Greenhouse Gas Inventories—Volume 4: Agriculture, Land Use and Forestry (GL-AFOLU). Available online: http://www.ipcc-nggip.iges.or.jp/public/2006gl/vol4.html (accessed on 10 December 2014).
- Lin, C.; Trianingsih, D. Identifying forest ecosystem regions for agricultural use and conservation. Sci. Agricola 2016, 73, 62–70. [Google Scholar] [CrossRef]
| Model ID | Formula (AGC Is in Ton per Tree) | R2 |
|---|---|---|
| C1 | AGC = 0.000022255 × LDBH2.2354 × LH0.5105 × LCI−0.0145 × LCR−0.1734 | 0.91 |
| C2 | AGC = 0.000068159 × LDBH1.4299 × LH1.1708 × LCI−0.0573 | 0.91 |
| C3 | AGC = 0.000016892 × LDBH2.4111 × LH0.3669 × LCR−0.1990 | 0.91 |
| C4 | AGC = 0.000031586 × LDBH1.8245 × LH0.8504 | 0.91 |
| C5 | AGC = 0.000013752 × LDBH2.6511 | 0.89 |
| C6 | AGC = 0.000475840 × LH2.4300 | 0.84 |
| C7 | AGC = exp(−7.5245 + 2.3949lnLH − 0.0145LCI + 0.0296LCR) | 0.91 |
| Model ID | Formula (Volume Is in the Unit of m3 per Tree) #,## | R2 |
|---|---|---|
| V1 | AGC = (a × LDBH1b × LHc) × D × BEF × CF | - |
| V2 | AGC = (a × LDBH2b × LHc) × D × BEF × CF | - |
| V3 | AGC = (a × LDBH3b × LHc) × D × BEF × CF | - |
| V4 | AGC = (a × LDBH4b × LHc) × D × BEF × CF | - |
| V5 | AGC = [exp(−6.2803 + 2.3774lnLH − 0.0145LCI + 0.0316LCR)] × D × BEF × CF | 0.91 |
| V6 | AGC = [exp(−6.1503 + 2.4012lnLH − 0.0165LCI)] × D × BEF × CF | 0.90 |
| V7 | AGC = [exp(−6.4149 + 2.4141lnLH)] × D × BEF × CF | 0.84 |
| V8 | AGC = [exp(−6.1846 + 2.3718lnLH − 0.0148LCI+0.0028LCR2)] × D × BEF × CF | 0.91 |
| V9 | AGC = [exp(−6.5963 + 2.3671lnLH + 0.0585LCR)] × D × BEF × CF | 0.86 |
| V1M | AGC = [0.2980(a × LDBH1b × LHc)1.0102] | 0.84 |
| V2M | AGC = [0.2929(a × LDBH2b × LHc)0.9933] | 0.86 |
| V3M | AGC = [0.3039(a × LDBH3b × LHc)1.0031] | 0.90 |
| V4M | AGC = [0.2919(a × LDBH4b × LHc)1.0026] | 0.91 |
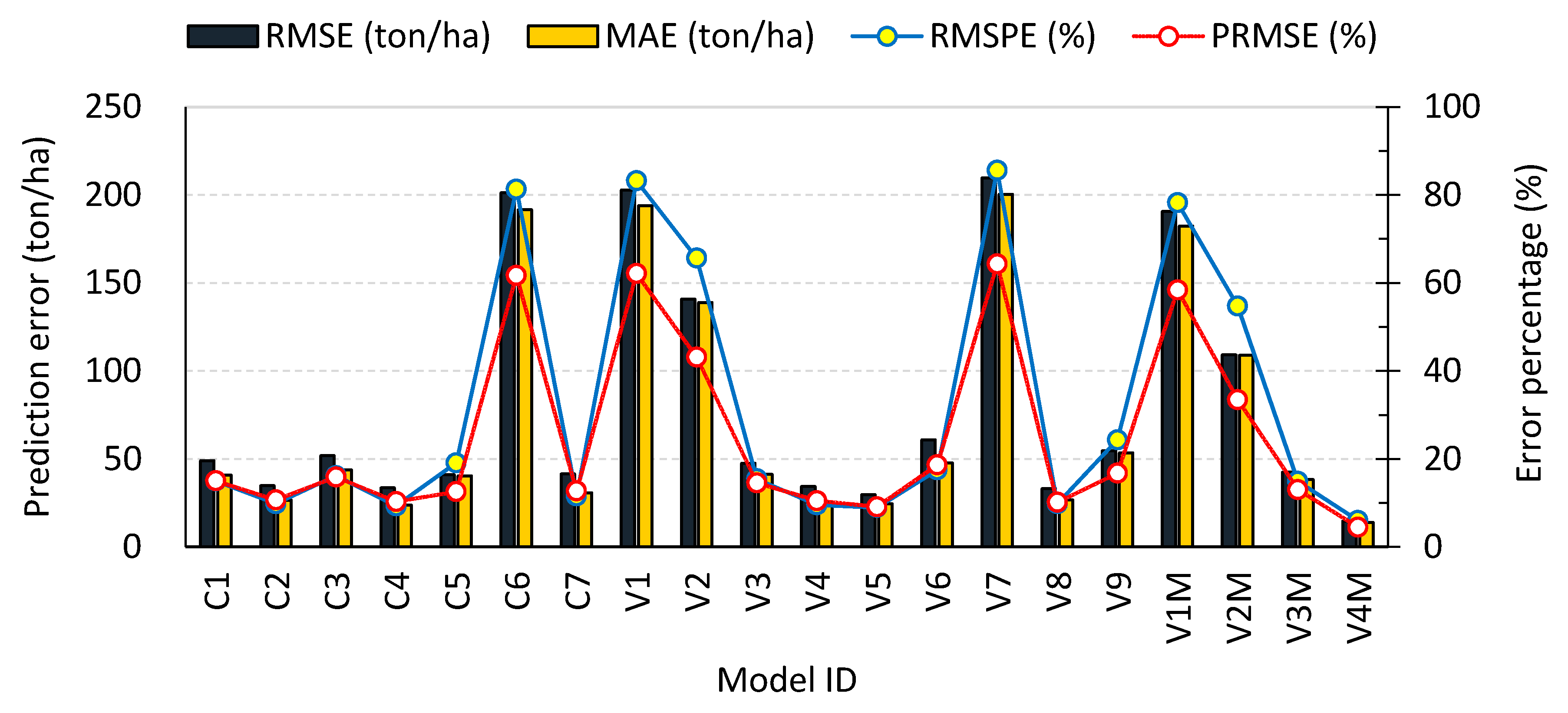
| Models ID | C1 | C2 | C3 | C4 | C5 | C6 | C7 | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V1M | V2M | V3M | V4M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAE (ton) | 0.17 | 0.18 | 0.17 | 0.17 | 0.18 | 0.21 | 0.18 | 0.30 | 0.21 | 0.20 | 0.19 | 0.18 | 0.19 | 0.21 | 0.18 | 0.19 | 0.21 | 0.20 | 0.19 | 0.17 |
| RMSE (ton) | 0.26 | 0.27 | 0.26 | 0.26 | 0.27 | 0.31 | 0.27 | 0.32 | 0.29 | 0.30 | 0.28 | 0.27 | 0.29 | 0.31 | 0.27 | 0.28 | 0.32 | 0.29 | 0.29 | 0.27 |
| PRMSE (%) | 32 | 33 | 32 | 32 | 33 | 38 | 33 | 38 | 35 | 36 | 34 | 33 | 35 | 38 | 32 | 34 | 38 | 35 | 35 | 32 |
| RMSPE (%) | 12 | 14 | 11 | 12 | 17 | 32 | 12 | 38 | 27 | 14 | 17 | 13 | 14 | 35 | 13 | 23 | 33 | 21 | 13 | 13 |
| Species | Statistics | AGC | C1 | C2 | C3 | C4 | C5 | C6 | C7 | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V1M | V2M | V3M | V4M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Japanese | Min | 0.57 | 0.01 | 0.04 | 0.01 | 0.02 | 0.00 | 0.34 | 0.02 | 0.35 | 0.31 | 0.00 | 0.02 | 0.01 | 0.01 | 0.36 | 0.01 | 0.20 | 0.34 | 0.29 | 0.00 | 0.02 |
| cedar | Max | 3.53 | 3.98 | 3.20 | 4.11 | 3.04 | 2.84 | 2.97 | 3.22 | 2.95 | 2.59 | 3.54 | 3.11 | 3.19 | 3.59 | 2.98 | 3.17 | 1.81 | 2.90 | 2.41 | 3.50 | 2.95 |
| Range | 2.96 | 3.98 | 3.16 | 4.10 | 3.01 | 2.83 | 2.63 | 3.20 | 2.59 | 2.29 | 3.54 | 3.10 | 3.18 | 3.59 | 2.63 | 3.16 | 1.61 | 2.56 | 2.12 | 3.49 | 2.93 | |
| SD | 0.56 | 0.91 | 0.77 | 0.93 | 0.77 | 0.73 | 0.61 | 0.82 | 0.60 | 0.53 | 0.92 | 0.79 | 0.81 | 0.93 | 0.61 | 0.82 | 0.37 | 0.59 | 0.49 | 0.91 | 0.75 | |
| Average | 1.26 | 1.47 | 1.43 | 1.48 | 1.42 | 1.18 | 1.95 | 1.45 | 1.95 | 1.71 | 1.47 | 1.42 | 1.41 | 1.52 | 1.97 | 1.42 | 1.13 | 1.91 | 1.60 | 1.44 | 1.35 | |
| Taiwan | Min | 0.14 | 0.01 | 0.03 | 0.01 | 0.02 | 0.01 | 0.27 | 0.02 | 0.28 | 0.25 | 0.01 | 0.02 | 0.01 | 0.01 | 0.29 | 0.01 | 0.16 | 0.27 | 0.23 | 0.01 | 0.02 |
| red | Max | 1.32 | 1.97 | 2.07 | 1.94 | 1.92 | 1.97 | 1.62 | 1.99 | 1.63 | 1.72 | 2.00 | 2.02 | 2.03 | 2.03 | 1.65 | 2.00 | 1.30 | 1.59 | 1.61 | 1.98 | 1.91 |
| cypress | Range | 1.18 | 1.96 | 2.04 | 1.93 | 1.90 | 1.97 | 1.35 | 1.97 | 1.34 | 1.47 | 2.00 | 2.00 | 2.02 | 2.02 | 1.36 | 1.98 | 1.14 | 1.32 | 1.37 | 1.97 | 1.89 |
| SD | 0.15 | 0.32 | 0.30 | 0.32 | 0.30 | 0.29 | 0.24 | 0.31 | 0.24 | 0.24 | 0.32 | 0.31 | 0.31 | 0.33 | 0.24 | 0.31 | 0.18 | 0.24 | 0.22 | 0.32 | 0.29 | |
| Average | 0.31 | 0.37 | 0.42 | 0.36 | 0.41 | 0.31 | 0.78 | 0.40 | 0.80 | 0.74 | 0.36 | 0.41 | 0.39 | 0.38 | 0.81 | 0.39 | 0.53 | 0.77 | 0.70 | 0.35 | 0.38 |
| Models ID | C1 | C2 | C3 | C4 | C5 | C6 | C7 | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V1M | V2M | V3M | V4M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | 91 | 91 | 91 | 91 | 89 | 84 | 91 | - | - | - | - | 91 | 90 | 84 | 91 | 86 | 84 | 86 | 90 | 91 |
| OPP | 85 | 90 | 84 | 90 | 84 | 29 | 88 | 27 | 46 | 85 | 90 | 91 | 82 | 25 | 90 | 79 | 32 | 56 | 86 | 95 |
| Uncertainty # | −6 | −1 | −7 | −1 | −5 | −55 | −3 | - | - | - | - | 0 | −8 | −59 | −1 | −7 | −52 | −30 | −4 | 4 |
| Model ID | V4M | V5 | C2 | V4 | V8 | C4 | C7 | V3M | C5 | V3 |
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (AIP) | 93 | 87 | 86 | 86 | 86 | 86 | 83 | 81 | 80 | 79 |
| Grouping | A | B | B, C | B, C | B, C | B, C | C, D | D, E | D, E | D, E |
| Model ID | C1 | C3 | V6 | V9 | V2M | V2 | V1M | C6 | V1 | V7 |
| Mean (AIP) | 79 | 77 | 74 | 73 | 44 | 30 | 9 | 4 | 3 | 0 |
| Grouping # | D, E | E, F | F, G | G | H | I | J | K | K, L | L |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Thomson, G.; Popescu, S.C. An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index. Remote Sens. 2016, 8, 528. https://doi.org/10.3390/rs8060528
Lin C, Thomson G, Popescu SC. An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index. Remote Sensing. 2016; 8(6):528. https://doi.org/10.3390/rs8060528
Chicago/Turabian StyleLin, Chinsu, Gavin Thomson, and Sorin C. Popescu. 2016. "An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index" Remote Sensing 8, no. 6: 528. https://doi.org/10.3390/rs8060528
APA StyleLin, C., Thomson, G., & Popescu, S. C. (2016). An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index. Remote Sensing, 8(6), 528. https://doi.org/10.3390/rs8060528