Satellite SST-Based Coral Disease Outbreak Predictions for the Hawaiian Archipelago
Abstract
:1. Introduction
2. Experimental Section
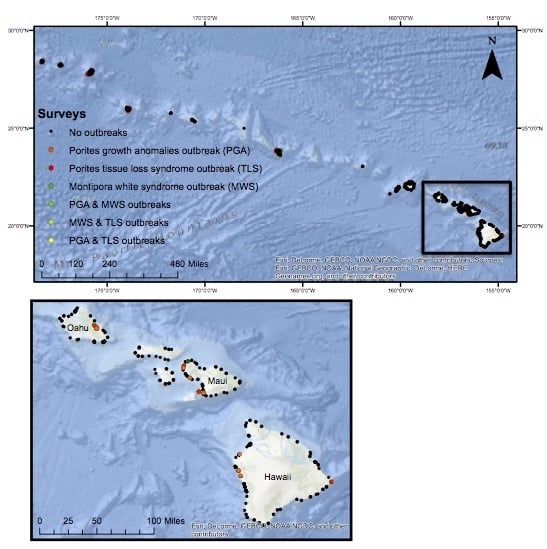
2.1. Field Surveys of Coral Diseases

| Variable | Type | Description and Unit | Min | Max |
|---|---|---|---|---|
| Total coral abundance | Biotic | Number of colonies/survey | 20 | 2633 |
| Total coral density | Biotic | Number of colonies/m2 | 0.15 | 52.4 |
| Porites density | Biotic | Number of colonies/m2 | 0.02 | 9.49 |
| Montipora density | Biotic | Number of colonies/m2 | 0.02 | 8.4 |
| Depth | Abiotic | Meters below sea surface | <1 | 24.5 |
| Winter Condition | Abiotic | Accumulation of positive and negative thermal anomalies; °C-weeks | −10.162 | 21.465 |
| Cold Snap | Abiotic | Magnitude and duration of cold stress; °C-weeks | −5.0255 | 0 |
| MPSA | Abiotic | Mean number of degree heating days in summer; °C | 0 | 0.78077 |
| Hot Snap | Abiotic | Magnitude and duration of heat stress; °C-weeks | 0 | 11.02 |
2.2. Defining Disease Outbreaks
2.3. SST-Based Metrics
2.4. Determining Outbreak Risk
3. Results and Discussion
3.1. Defining Disease Outbreaks

3.2. Determining Outbreak Risk
| Coral Disease | Model | nt | tc | lr | bf | cv dev | se | AUC | D |
|---|---|---|---|---|---|---|---|---|---|
| Montipora white syndrome | PA | 3500 | 5 | 0.001 | 0.75 | 0.262 | 0.036 | 0.70 | 0.30 |
| PIP | 1400 | 4 | 0.001 | 0.75 | 0.113 | 0.086 | |||
| Porites growth anomalies | PA | 1350 | 4 | 0.005 | 0.75 | 1.061 | 0.031 | 0.85 | 0.41 |
| PIP | 1900 | 5 | 0.005 | 0.75 | 0.048 | 0.005 | |||
| Porites tissue loss | PA | 750 | 3 | 0.005 | 0.75 | 1.213 | 0.027 | 0.67 | 0.44 |
| PIP | 2700 | 4 | 0.005 | 0.75 | 0.33 | 0.004 |
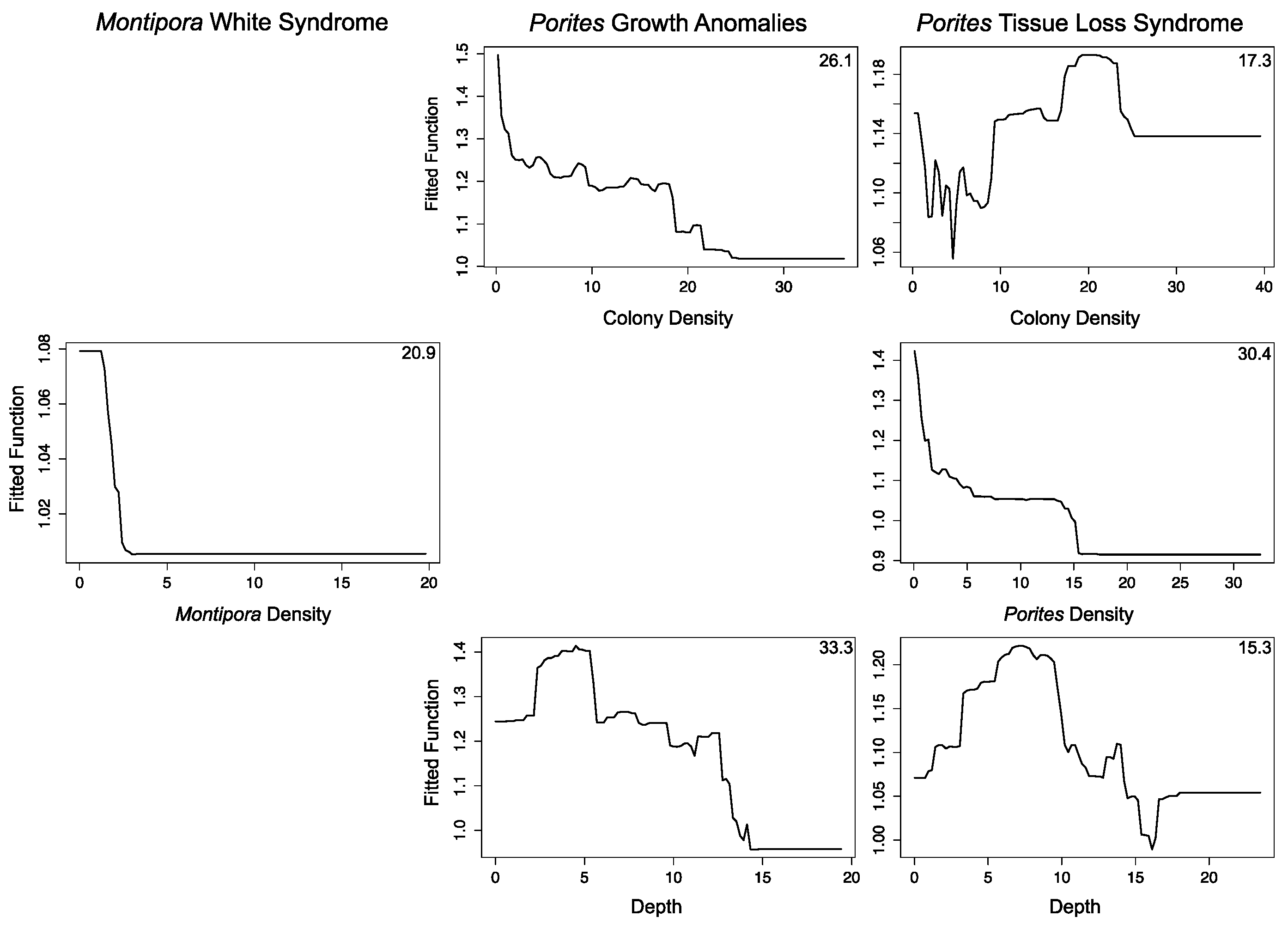
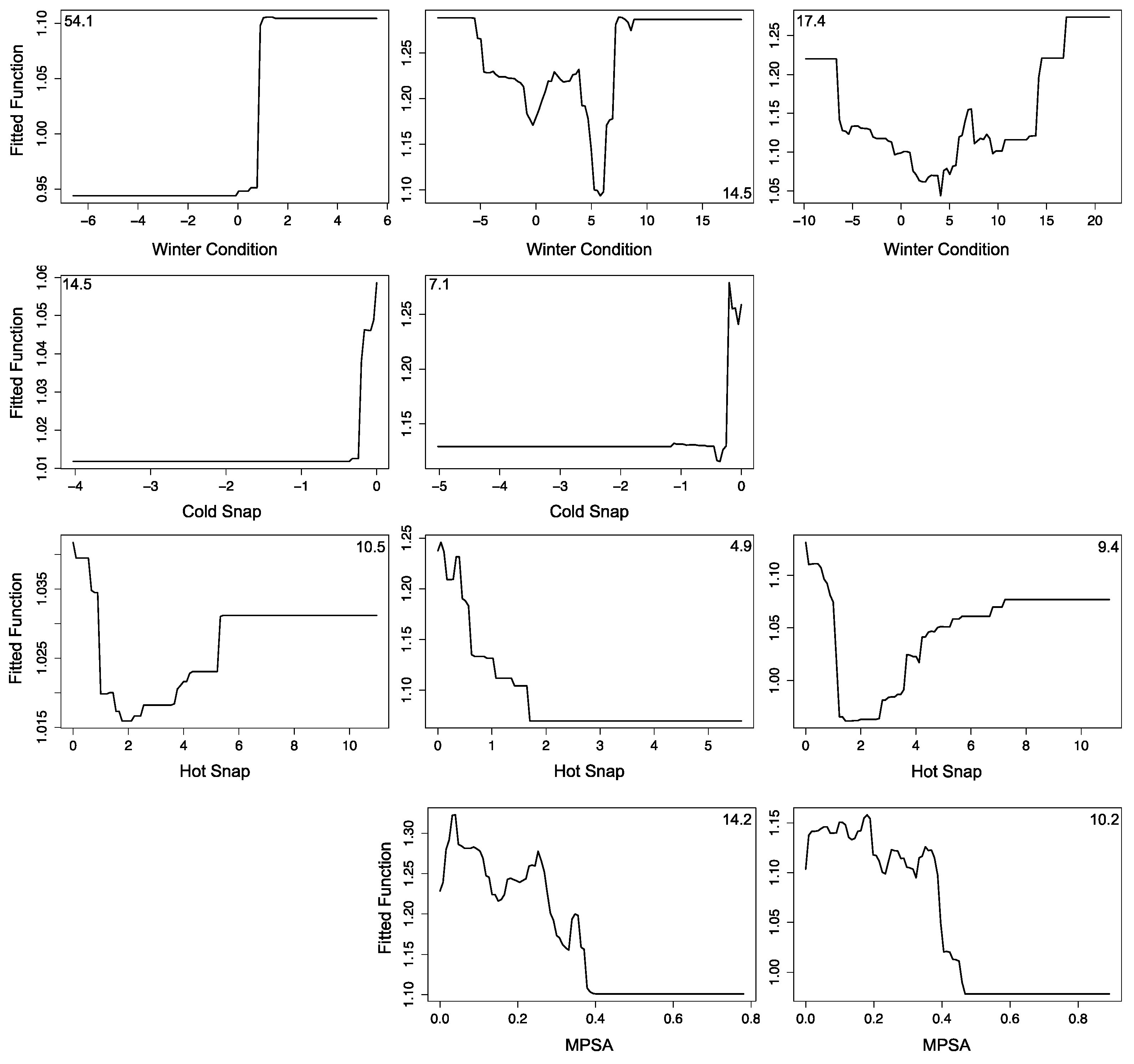
3.2.1. Montipora White Syndrome
3.2.2. Porites Growth Anomalies
3.2.3. Porites Tissue Loss Syndrome
3.3. Forecasting Disease Risk
3.4. Uncertainties, Errors and Accuracies
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Harvell, C.D.; Mitchell, C.E.; Ward, J.R.; Altizer, S.; Dobson, A.P.; Ostfeld, R.S.; Samuel, M.D. Climate warming and disease risks for terrestrial and marine biota. Sci. Rev. Ecol. 2002, 296, 2158–2162. [Google Scholar] [CrossRef] [PubMed]
- Burge, C.A.; Eakin, C.M.; Friedman, C.S.; Froelich, B.; Hershberger, P.K.; Hofmann, E.E.; Petes, L.E.; Prager, K.C.; Weil, E.; Willis, B.L.; et al. Climate change influences on marine infectious diseases: Implications for management and society. Ann. Rev. Mar. Sci. 2014, 6, 249–277. [Google Scholar] [CrossRef] [PubMed]
- Vredenburg, V.T.; Knapp, R.A.; Tunstall, T.S.; Briggs, C.J. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc. Natl. Acad. Sci. USA 2010, 107, 9689–9694. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Altizer, S.; Dobson, A.; Hosseini, P.; Hudson, P.; Pascual, M.; Rohani, P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006, 9, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Travers, M.A.; Basuyaux, O.; le Goic, N.; Huchette, S.; Nicolas, J.L.; Koken, M.; Paillard, C. Influence of temperature and spawning effort on Haliotis tuberculata mortalities caused by Vibrio harveyi: An example of emerging vibriosis linked to global warming. Glob. Chang. Biol. 2009, 15, 1365–1376. [Google Scholar] [CrossRef]
- Carpenter, K.E.; Abrar, M.; Aeby, G.; Aronson, R.B.; Banks, S.; Bruckner, A.; Chiriboga, A.; Cortés, J.; Delbeek, J.C.; Devantier, L.; et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 2008, 321, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Reytar, K.; Spalding, M.; Perry, A. Reefs at Risk Revisited; World Resources Institute: Washington, DC, USA, 2011. [Google Scholar]
- Ruiz-Moreno, D.; Willis, B.L.; Page, A.C.; Weil, E.; Cróquer, A.; Vargas-Angel, B.; Jordan-Garza, A.G.; Jordán-Dahlgren, E.; Raymundo, L.; Harvell, C.D. Global coral disease prevalence associated with sea temperature anomalies and local factors. Dis. Aquat. Org. 2012, 100, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.W.; Richardson, L.L. Emerging coral diseases: A temperature-driven process? Mar. Ecol. 2014, 36, 1–14. [Google Scholar] [CrossRef]
- Bruckner, A.W.; Hill, R.L. Ten years of change to coral comunities off Mona and Desecheo Islands, Puerto Rico, from disease and bleaching. Dis. Aquat. Org. 2009, 87, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Sokolow, S. Effects of a changing climate on the dynamics of coral infectious disease: A review of the evidence. Dis. Aquat. Org. 2009, 87, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.R.; Kim, K.; Harvell, C.D. Temperature affects coral disease resistance and pathogen growth. Mar. Ecol. Prog. Ser. 2007, 329, 115–121. [Google Scholar] [CrossRef]
- Aeby, G.S. Outbreak of coral disease in the Northwestern Hawaiian Islands. Coral Reefs 2005, 24, 481. [Google Scholar] [CrossRef]
- Hobbs, J.P.A.; Frisch, A.J.; Newman, S.J.; Wakefield, C.B. Selective Impact of Disease on Coral Communities: Outbreak of White Syndrome Causes Significant Total Mortality of Acropora Plate Corals. PLoS ONE 2015, 10, e0132528. [Google Scholar]
- Williams, G.J.; Knapp, I.S.; Work, T.M.; Conklin, E.J. Outbreak of Acropora white syndrome following a mild bleaching event at Palmyra Atoll, Northern Line Islands, Central Pacific. Coral Reefs 2011, 30, 621. [Google Scholar] [CrossRef]
- Heron, S.F.; Willis, B.L.; Skirving, W.J.; Eakin, C.M.; Page, C.A.; Miller, I.R. Summer hot snaps and winter conditions: Modelling white syndrome outbreaks on Great Barrier Reef corals. PLoS ONE 2010, 5, e12210. [Google Scholar] [CrossRef] [PubMed]
- Maynard, J.A.; Anthony, K.R.N.; Harvell, C.D.; Burgman, M.A.; Beeden, R.; Sweatman, H.; Heron, S.F.; Lamb, J.B.; Willis, B.L. Predicting outbreaks of a climate-driven coral disease in the Great Barrier Reef. Coral Reefs 2011, 30, 485–495. [Google Scholar] [CrossRef]
- Eakin, C.M.; Morgan, J.A.; Heron, S.F.; Smith, T.B.; Liu, G.; Alvarez-Filip, L.; Baca, B.; Bartels, E.; Bastidas, C.; Bouchon, C.; et al. Caribbean corals in crisis: Record thermal stress, bleaching, and mortality in 2005. PLoS ONE 2010, 5, e13969. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.F.; Selig, E.R.; Casey, K.S.; Page, C.A.; Willis, B.L.; Harvell, C.D.; Sweatman, H.; Melendy, A.M. Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 2007, 5, e124. [Google Scholar] [CrossRef] [PubMed]
- Experimental Coral Disease Outbreak RiskGreat Barrier Reef, Australia. Available online: coralreefwatch.noaa.gov/satellite/disease/dz_gbr.php (accessed on 15 September 2015).
- Randall, C.J.; van Woesik, R. Contemporary white-band disease in Caribbean corals driven by climate change. Nat. Clim. Chang. 2015, 5, 375–379. [Google Scholar] [CrossRef]
- Beeden, R.; Maynard, J.A.; Marshall, P.A.; Heron, S.F.; Willis, B.L. A framework for responding to coral disease outbreaks that facilitates adaptive management. Environ. Manag. 2012, 49, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Aeby, G.S.; Ross, M.; Williams, G.J.; Lewis, T.D.; Work, T.M. Disease dynamics of Montipora white syndrome within Kaneohe Bay, Oahu, Hawai’i: Distribution, seasonality, virulence, and transmissibility. Dis. Aquat. Org. 2010, 91, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Work, T.M.; Russell, R.; Aeby, G.S. Tissue loss (white syndrome) in the coral Montipora capitata is a dynamic disease with multiple host responses and potential causes. Proc. R. Soc. B Biol. Sci. 2012, 279, 4334–4341. [Google Scholar] [CrossRef] [PubMed]
- Aeby, G.S.; Williams, G.J.; Franklin, E.C.; Kenyon, J.; Cox, E.F.; Coles, S.; Work, T.M. Patterns of Coral Disease across the Hawaiian Archipelago: Relating Disease to Environment. PLoS ONE 2011, 6, e20370. [Google Scholar] [CrossRef] [PubMed]
- Stimson, J. Ecological characterization of coral growth anomalies on Porites compressa in Hawai’i. Coral Reefs 2010, 30, 133–142. [Google Scholar] [CrossRef]
- Williams, G.J.; Aeby, G.S.; Cowie, R.O.M.; Davy, S.K. Predictive modeling of coral disease distribution within a reef system. PLoS ONE 2010, 5, e9264. [Google Scholar] [CrossRef] [PubMed]
- Kimes, N.E.; Grim, C.J.; Johnson, W.R.; Hasan, N.A.; Tall, B.D.; Kothary, M.H.; Kiss, H.; Munk, A.C.; Tapia, R.; Green, L.; et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012, 6, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, B.; Videau, P.; Burger, A.; Shore-Maggio, A.; Runyon, C.M.; Sudek, M.; Aeby, G.S.; Callahan, S.M. Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl. Environ. Microbiol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Casey, K.S.; Brandon, T.B.; Comillon, P.; Evans, R. The Path, Present, and Future of the AVHRR Pathfinder SST Program. Oceanogr. Space 2010, 5, 273–287. [Google Scholar]
- Liu, G.; Heron, S.; Eakin, C.; Muller-Karger, F.; Vega-Rodriguez, M.; Guild, L.; de la Cour, J.; Geiger, E.; Skirving, W.; Burgess, T.; et al. Reef-Scale Thermal Stress Monitoring of Coral Ecosystems: New 5-km Global Products from NOAA Coral Reef Watch. Remote Sens. 2014, 6, 11579–11606. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Cappelle, J.; Girard, O.; Fofana, B.; Gaidet, N.; Gilbert, M. Ecological Modeling of the Spatial Distribution of Wild Waterbirds to Identify the Main Areas Where Avian Influenza Viruses are Circulating in the Inner Niger Delta, Mali. Ecohealth 2010, 7, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Franklin, E.C.; Jokiel, P.L.; Donahue, M.J. Predictive modeling of coral distribution and abundance in the Hawaiian Islands. Mar. Ecol. Prog. Ser. 2013, 481, 121–132. [Google Scholar] [CrossRef]
- R Core Team: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing). 2014. Available online: http://www.r-project.org (accessed on 15 December 2015).
- Aeby, G.S. Baseline Levels of Coral Disease in the Northwestern Hawaiian Islands. Available online: http://www.biodiversitylibrary.org/part/83092#/summary (accessed on 15 December 2015).
- Aeby, G.S.; Williams, G.J.; Franklin, E.C.; Haapkyla, J.; Harvell, C.D.; Neale, S.; Page, C.A.; Raymundo, L.; Vargas-Ángel, B.; Willis, B.L.; et al. Growth anomalies on the coral genera Acropora and Porites are strongly associated with host density and human population size across the Indo-Pacific. PLoS ONE 2011, 6, e16887. [Google Scholar] [CrossRef] [PubMed]
- Couch, C.S.; Garriques, J.D.; Barnett, C.; Preskitt, L.; Cotton, S.; Giddens, J.; Walsh, W. Spatial and temporal patterns of coral health and disease along leeward Hawai’i Island. Coral Reefs 2014, 33, 693–704. [Google Scholar] [CrossRef]
- Kermack, W.O.; McKendrick, A.G. A Contribution to the Mathematical Theory of Epidemics. Proc. R. Soc. A Math. Phys. Eng. Sci. 1927, 115, 700–721. [Google Scholar] [CrossRef]
- Lloyd-Smith, J.O.; Cross, P.C.; Briggs, C.J.; Daugherty, M.; Getz, W.M.; Latto, J.; Sanchez, M.S.; Smith, A.B.; Swei, A. Should we expect population thresholds for wildlife disease? Trends Ecol. E 2005, 20, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Hawley, D.M.; Altizer, S.M. Disease ecology meets ecological immunology: Understanding the links between organismal immunity and infection dynamics in natural populations. Funct. Ecol. 2011, 25, 48–60. [Google Scholar] [CrossRef]
- Dube, D.; Kim, K.; Alker, A.P.; Harvell, C.D. Size structure and geographic variation in chemical resistance of sea fan corals Gorgonia ventalina to a fungal pathogen. Mar. Ecol. Prog. Ser. 2002, 231, 139–150. [Google Scholar] [CrossRef]
- Vega-Thurber, R.L.; Burkepile, D.E.; Fuchs, C.; Shantz, A.A.; McMinds, R.; Zaneveld, J.R. Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Chang. Biol. 2014, 20, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Haapkyla, J.; Unsworth, R.K.F.; Flavell, M.; Bourne, D.G.; Schaffelke, B.; Willis, B.L. Seasonal Rainfall and Runoff Promote Coral Disease on an Inshore Reef. PLoS ONE 2011, 6, e16893. [Google Scholar] [CrossRef]
- Ban, S.S.; Graham, N.A.J.; Connolly, S.R. Evidence for multiple stressor interactions and effects on coral reefs. Glob. Chang. Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Gove, J.M.; Williams, G.J.; McManus, M.A.; Heron, S.F.; Sandin, S.A.; Vetter, O.J.; Foley, D.G. Quantifying climatological ranges and anomalies for pacific coral reef ecosystems. PLoS ONE 2013, 8, e61974. [Google Scholar] [CrossRef] [PubMed]
- Jokiel, P.L.; Brown, E.K. Global warming, regional trends and inshore environmental conditions influence coral bleaching in Hawaii. Glob. Chang. Biol. 2004, 10, 1627–1641. [Google Scholar] [CrossRef]
- NOAA Coral Reef Watch—2014 Annual Summaries of Thermal Conditions Related to Coral Bleaching for U.S. National Coral Reef Monitoring Program (NCRMP) Jurisdictions. 2014. Available online: http://coralreefwatch.noaa.gov/satellite/analyses_guidance/2014_annual_summaries_thermal_stress_conditions_NCRMP.pdf (accessed on 15 December 2015).
- Department of Land and Natural Resources. Record Ocean Temperatures Causing Coral Bleaching across Hawaii. 2015. Available online: http://dlnr.hawaii.gov/blog/2015/09/11/nr15-135/ (accessed on 15 December 2015). [Google Scholar]
- Experimental Coral Disease Outbreak RiskHawaiian Archipelago. Available online: http://coralreefwatch.noaa.gov/satellite/disease_haw/dz_hawaii.php (accessed on 15 September 2015).
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caldwell, J.M.; Heron, S.F.; Eakin, C.M.; Donahue, M.J. Satellite SST-Based Coral Disease Outbreak Predictions for the Hawaiian Archipelago. Remote Sens. 2016, 8, 93. https://doi.org/10.3390/rs8020093
Caldwell JM, Heron SF, Eakin CM, Donahue MJ. Satellite SST-Based Coral Disease Outbreak Predictions for the Hawaiian Archipelago. Remote Sensing. 2016; 8(2):93. https://doi.org/10.3390/rs8020093
Chicago/Turabian StyleCaldwell, Jamie M., Scott F. Heron, C. Mark Eakin, and Megan J. Donahue. 2016. "Satellite SST-Based Coral Disease Outbreak Predictions for the Hawaiian Archipelago" Remote Sensing 8, no. 2: 93. https://doi.org/10.3390/rs8020093