Self-Guided Segmentation and Classification of Multi-Temporal Landsat 8 Images for Crop Type Mapping in Southeastern Brazil
Abstract
:1. Introduction
2. Material
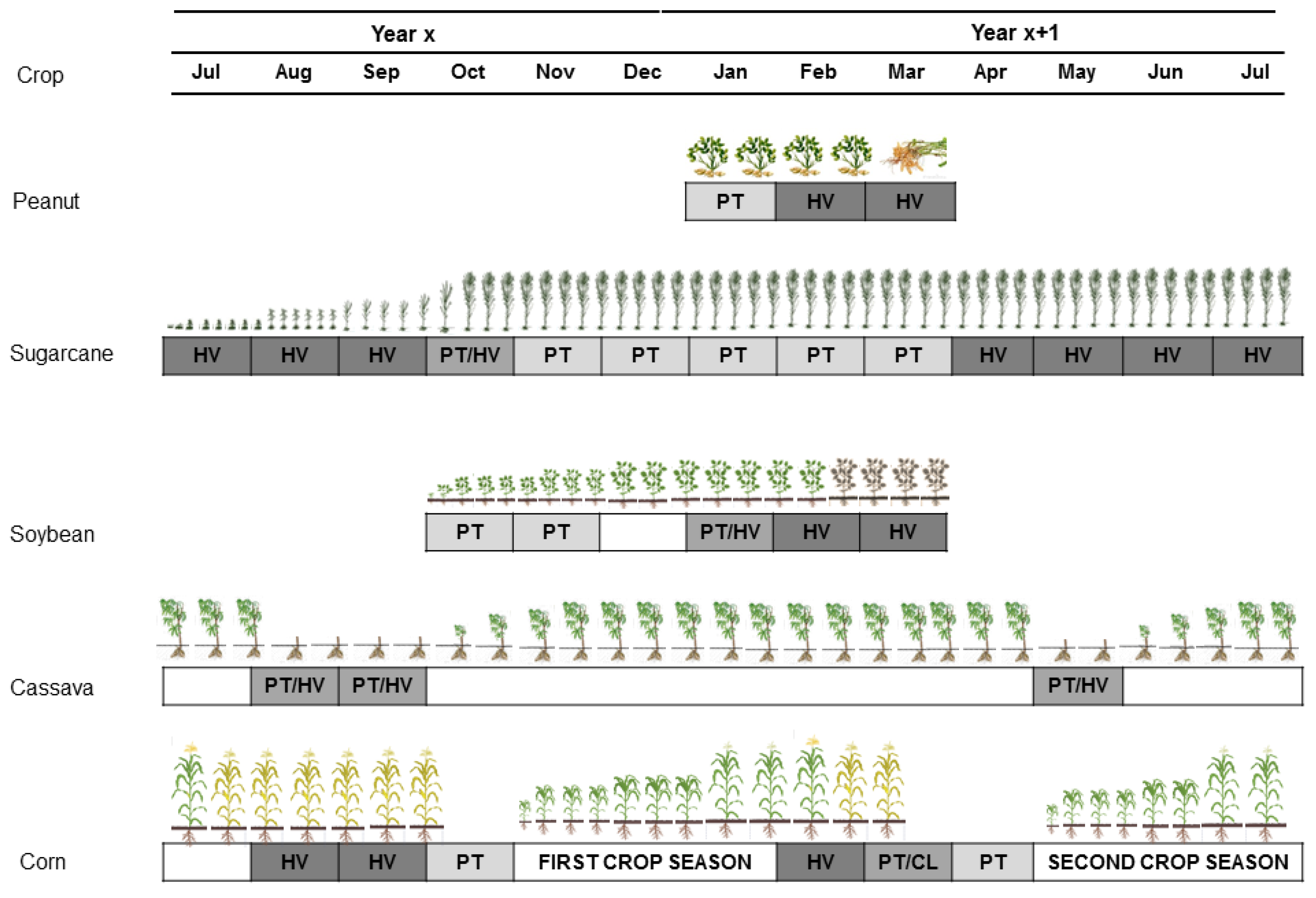
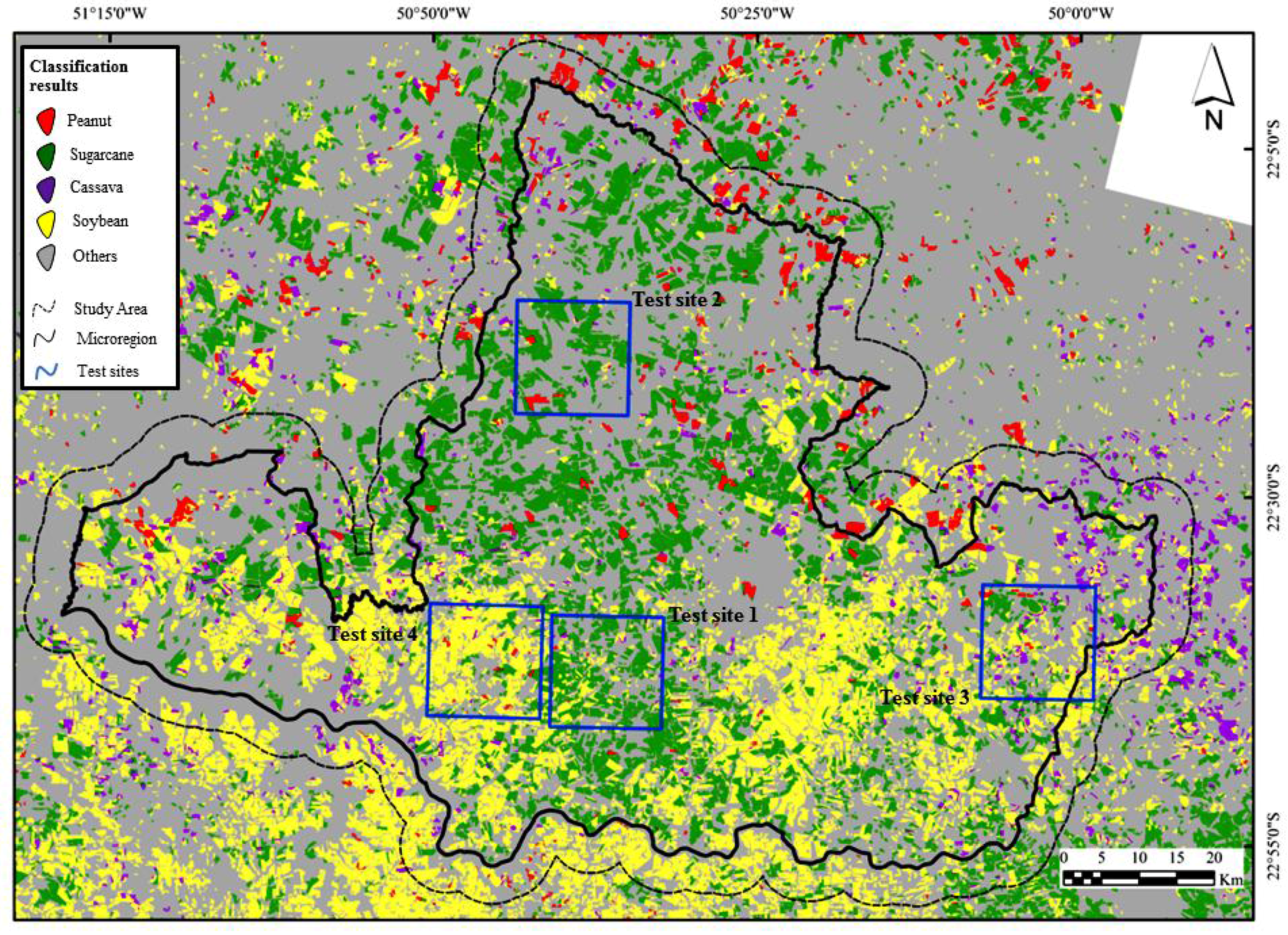
2.1. Study Site and Crop Calendar


2.2. Satellite Imagery
- Two multi-spectral Landsat 8 image mosaics were used for image segmentation and classification. Seven additional mosaics from Landsat 7 (Enhanced Thematic Mapper Plus, ETM+) and Landsat 8 (OLI) images were used for the generation of reference information.
- One RapidEye mosaic from four images was used as ancillary data for assisting in the visual photo-interpretation and for (manual) field delineation as suggested by Forster et al. [44].
| Images Description | Landsat 8 | Landsat 7 | RapidEye |
|---|---|---|---|
| Sensor name | Operator Land Imager (OLI) | Enhanced Thematic Mapper (ETM+) | Multispectral Imager (MSI) |
| Satellite (number) | 1 | 1 | 5 |
| Equator Time (hour) | ±11:00 | ±10:10 | ±10:30 |
| Radiometric resolution (Bits) | 16 | 8 | 12 |
| Ground Sampling Distance (m) | 30 | 30 | 6.5 |
| Spectral bands (nm) | |||
| Aerosols | 430–450 | -- | -- |
| Blue | 450–520 | 450–520 | 440–510 |
| Green | 530–590 | 530–610 | 520–590 |
| Red | 640–670 | 630–690 | 630–685 |
| Red-Edge | -- | -- | 690–730 |
| NIR | 850–880 | 780–900 | 760–850 |
| SWIR | 1570–1610 | 1550–1750 | -- |
| 2110–2290 | 2090–2350 | -- | |
| Panchromatic (PAN) | 500–680 | 520–900 | -- |
| Cirrus | 1360–1380 | -- | -- |
- A late-season mosaic combining scenes from 21 August 2013 and 30 August 2013,
- A mid-season mosaic using scenes from 28 January 2014 and 6 February 2014.
- Three mosaics between the late-season and mid-season (from October, November and December), and
- Four mosaics after mid-season (from February, March, April and May)
| Image Identifier | Landsat 8 and 7 (Path/Row) | RapidEye (Path/Row) | |||||
|---|---|---|---|---|---|---|---|
| 222/75 | 222/76 | 221/76 | 285/15 | 286/15 | 287/15 | 288/15 | |
| 16/17/18 | 16/17/18 | 16/17/18 | 16/17/18 | ||||
| Images for manual field delineation | 21 Aug. 2013 | 21 Aug. 2013 | 30 Aug. 2013 | 03 Jul. 2012 | 03 Jul. 2012 | 03 Jul. 2012 | 27 Jul. 2012 |
| 28 Jan. 2014 | 28 Jan. 2014 | 06 Feb. 2014 | |||||
| Images for visual photo-interpretation | 21 Aug. 2013 | 21.Aug.13 | 30 Aug. 2013 | -- | -- | -- | -- |
| 08 Oct. 2013 | 08 Oct. 2013 | 09 Oct. 2013 | -- | -- | -- | -- | |
| 09 Nov. 2013 | 09 Nov. 2013 | 02 Nov. 2013 | -- | -- | -- | -- | |
| 11 Dec. 2013 | 11 Dec. 2013 | 04 Dec. 2013 | -- | -- | -- | -- | |
| 28 Jan. 2014 | 28 Jan. 2014 | 06 Feb. 2014 | -- | -- | -- | -- | |
| 13 Feb. 2013 | 13 Feb. 2013 | 14 Feb. 2014 | -- | -- | -- | -- | |
| 01 Mar. 2013 | 01 Mar. 2013 | 10 Mar. 2013 | -- | -- | -- | -- | |
| 02 Apr. 2013 | 02 Apr 2013 | 11 Apr. 2013 | -- | -- | -- | -- | |
| 04 May 2013 | 04 May. 2013 | 13 May 2013 | -- | -- | -- | -- | |
| Images for segmentation and classification | 21 Aug. 2013 | 21 Aug. 2013 | 30 Aug. 2013 | -- | -- | -- | -- |
| 28 Jan. 2014 | 28 Jan. 2014 | 06 Feb. 2014 | -- | -- | -- | -- | |
| Total number of consulted images | 9 | 9 | 9 | 1 | 1 | 1 | 1 |
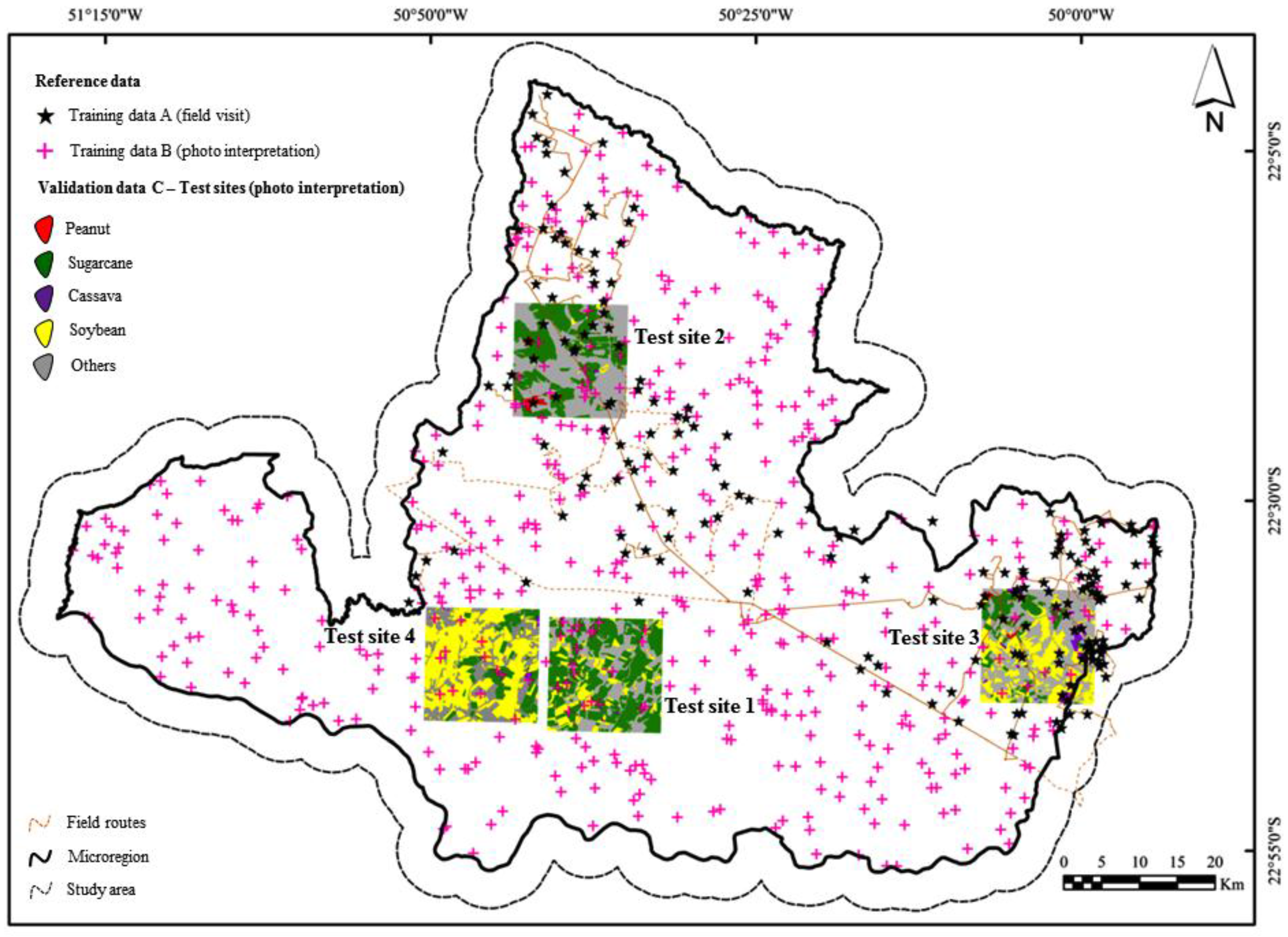
2.3. Reference Data Collection
- (A)
- Reference samples from field visit (n=177),
- (B)
- Reference samples obtained through manual photo interpretation (n=500), and
- (C)
- Manually delineated and interpreted fields (n=2502).
| Reference data | Data type | Interpretation | Peanut | Soybean | Cassava | Sugarcane | Others | Total |
|---|---|---|---|---|---|---|---|---|
| Training samples (A) | Points | Field visit | 24 | 33 | 37 | 32 | 51 | 177 |
| Training samples (B) | Points | Photo interpretation | 11 | 92 | 17 | 96 | 284 | 500 |
| Validation samples—test sites (C) | Polygons | Photo interpretation | 9 | 689 | 33 | 495 | 1275 | 2502 |
| Test Site | Metrics | Crops (area in ha) | Others | Average & (total) | |||
|---|---|---|---|---|---|---|---|
| Peanut | Soybean | Cassava | Sugarcane | ||||
| TS 1 | Average | 4.67 | 26.30 | 8.01 | 40.80 | 29.28 | 33.39 |
| Stand. Dev. | -- | 24.28 | 3.55 | 50.81 | 40.12 | 28.58 | |
| Nº Parcels | 1 | 126 | 7 | 288 | 252 | (n = 674) | |
| TS 2 | Average | 76.00 | 17.66 | 23.95 | 108.60 | 45.01 | 54.24 |
| Stand. Dev. | 46.48 | 16.01 | -- | 108.77 | 68.34 | 59.03 | |
| N° Parcels | 6 | 13 | 1 | 81 | 289 | (n = 390) | |
| TS 3 | Average | 62.94 | 30.86 | 21.81 | 53.76 | 26.07 | 29.64 |
| Stand. Dev. | 59.78 | 38.74 | 22.65 | 70.40 | 42.63 | 44.66 | |
| N° Parcels | 2 | 219 | 17 | 60 | 461 | (n = 759) | |
| TS 4 | Average | -- | 38.86 | 17.51 | 44.79 | 23.80 | 33.14 |
| Stand. Dev. | -- | 37.35 | 13.55 | 46.67 | 32.78 | 37.30 | |
| N° Parcels | 0 | 331 | 8 | 67 | 273 | (n = 679) | |
| all | Average | 65.17 | 33.62 | 17.91 | 53.98 | 30.51 | 35.97 |
| Stand. Dev. | 48.41 | 35.87 | 18.16 | 69.93 | 48.18 | 51.03 | |
| N° Parcels | 9 | 689 | 33 | 496 | 1275 | (n = 2502) | |
3. Methods
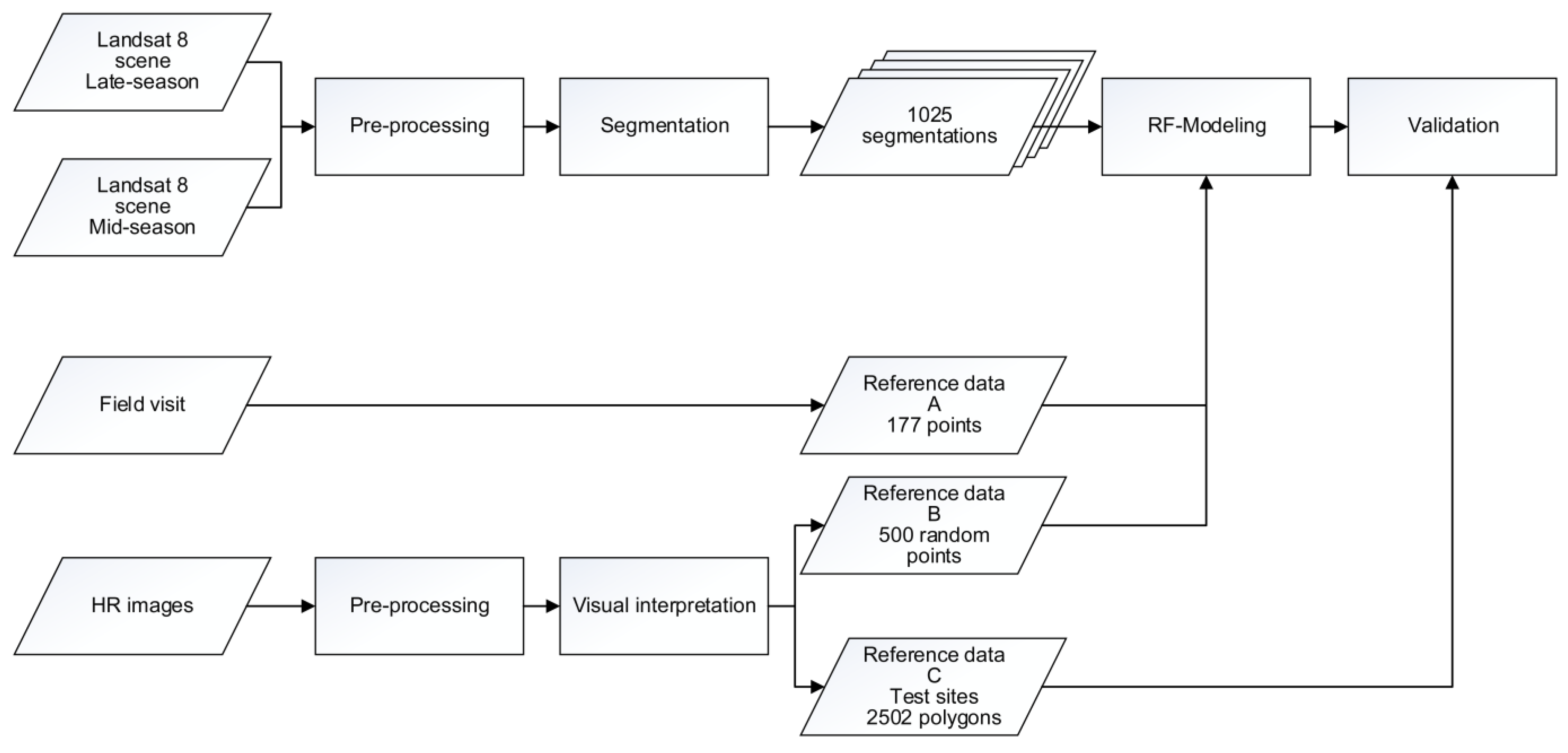
3.1. Image Segmentation and Data Extraction
- (1)
- The mean value and standard deviation of each spectral band for each of the two dates (6 × 2 × 2 = 24 variables);
- (2)
- Maximum difference and brightness (2 variables);
- (3)
- The normalized difference vegetation index (NDVI) for each date (2 variables);
- (4)
- The maximum, medium and minimum mode of the segments in each spectral band for each date (3 × 6 × 2 =36 variables);
- (5)
- The quartiles to the segments in each spectral band and each date (5 × 6 × 2 = 60 variables);
- (6)
- The GLCM (Gray-Level Co-Occurrence Matrix) homogeneity, GLCM contrast, GLCM dissimilarity, GLCM entropy, GLCM angular 2nd moment, GLCM mean, GLCM standard deviation, GLCM correlation for all directions (8 variables); and
- (7)
- Shape and size information of the shapes (17 variables).
3.2. Classification and Feature Selection
- For mtry (the number of input variables) the default setting was kept (i.e., the square root of the number of input variables).
- Ntree (the number of trees) was fixed to 1000.
3.3. Model Performance and Model Evaluation
- Based on the out-of-bag (OOB) samples from the (point) reference data set (i.e., samples A and B—see Table 3)
4. Results and Discussion
4.1. Model Accuracy OOB

| Min | Median | Max | |
|---|---|---|---|
| OA | 0.658 | 0.818 | 0.858 |
| Kappa | 0.476 | 0.722 | 0.786 |
| Producer’s Accuracy | User’s Accuracy | |||||
|---|---|---|---|---|---|---|
| min | median | max | min | Median | max | |
| Peanut | 0.379 | 0.743 | 0.879 | 0.550 | 0.931 | 1.000 |
| Sugarcane | 0.391 | 0.760 | 0.897 | 0.561 | 0.819 | 0.887 |
| Cassava | 0.000 | 0.273 | 0.537 | 0.000 | 0.632 | 0.867 |
| Soybean | 0.818 | 0.925 | 0.957 | 0.721 | 0.853 | 0.909 |
| Others | 0.583 | 0.803 | 0.874 | 0.549 | 0.723 | 0.786 |
| Peanut | Sugarcane | Cassava | Soybean | Others | Sum | User’s Accuracy | |
|---|---|---|---|---|---|---|---|
| Peanut | 26 | 1 | 0 | 0 | 0 | 27 | 0.963 |
| Sugarcane | 1 | 109 | 2 | 2 | 12 | 126 | 0.865 |
| Cassava | 0 | 0 | 20 | 5 | 0 | 25 | 0.800 |
| Soybean | 7 | 1 | 24 | 110 | 7 | 149 | 0.738 |
| Others | 1 | 14 | 8 | 11 | 316 | 350 | 0.903 |
| Sum | 35 | 125 | 54 | 128 | 335 | 677 | |
| Producer’s accuracy. | 0.743 | 0.872 | 0.370 | 0.859 | 0.943 | ||
| Overall accuracy (Kappa) | 0.858 (0.786) |
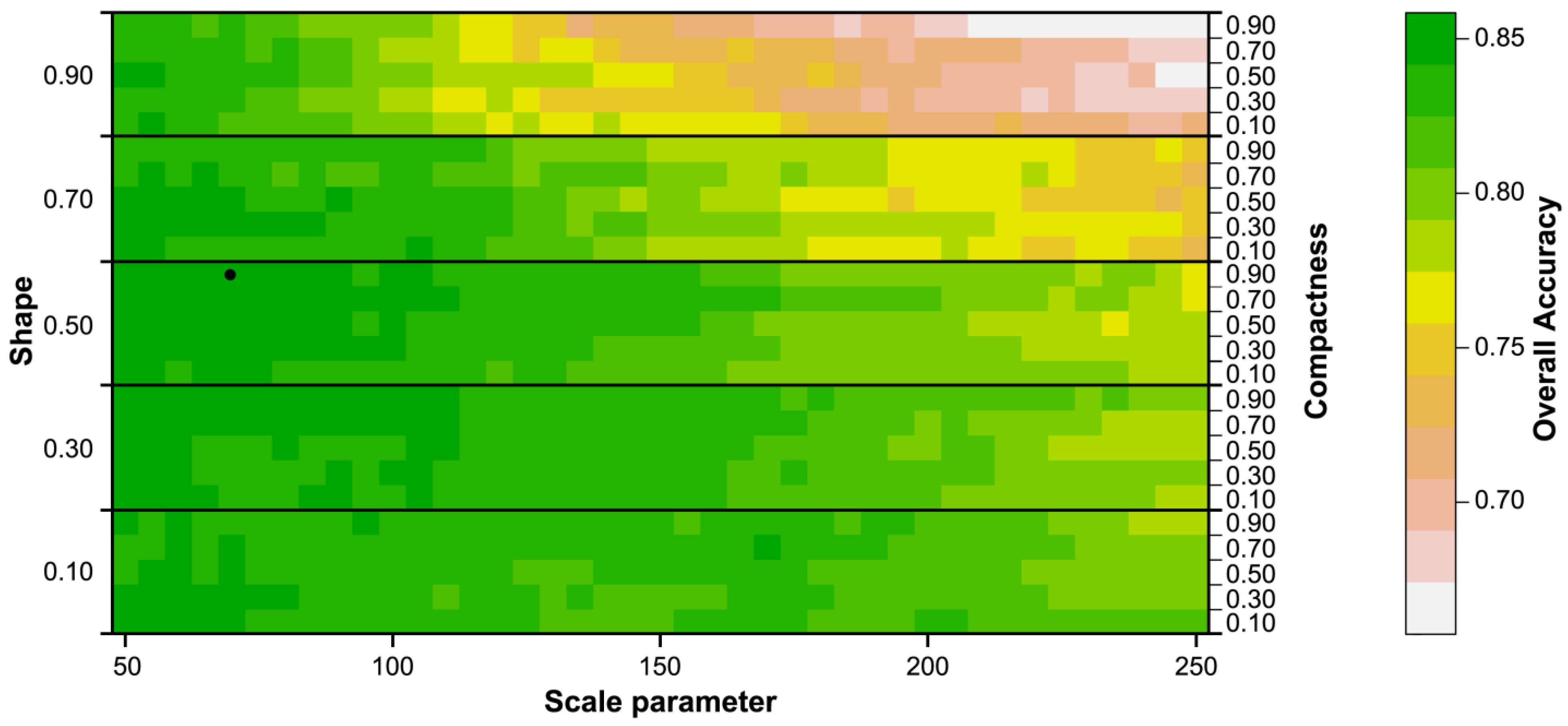
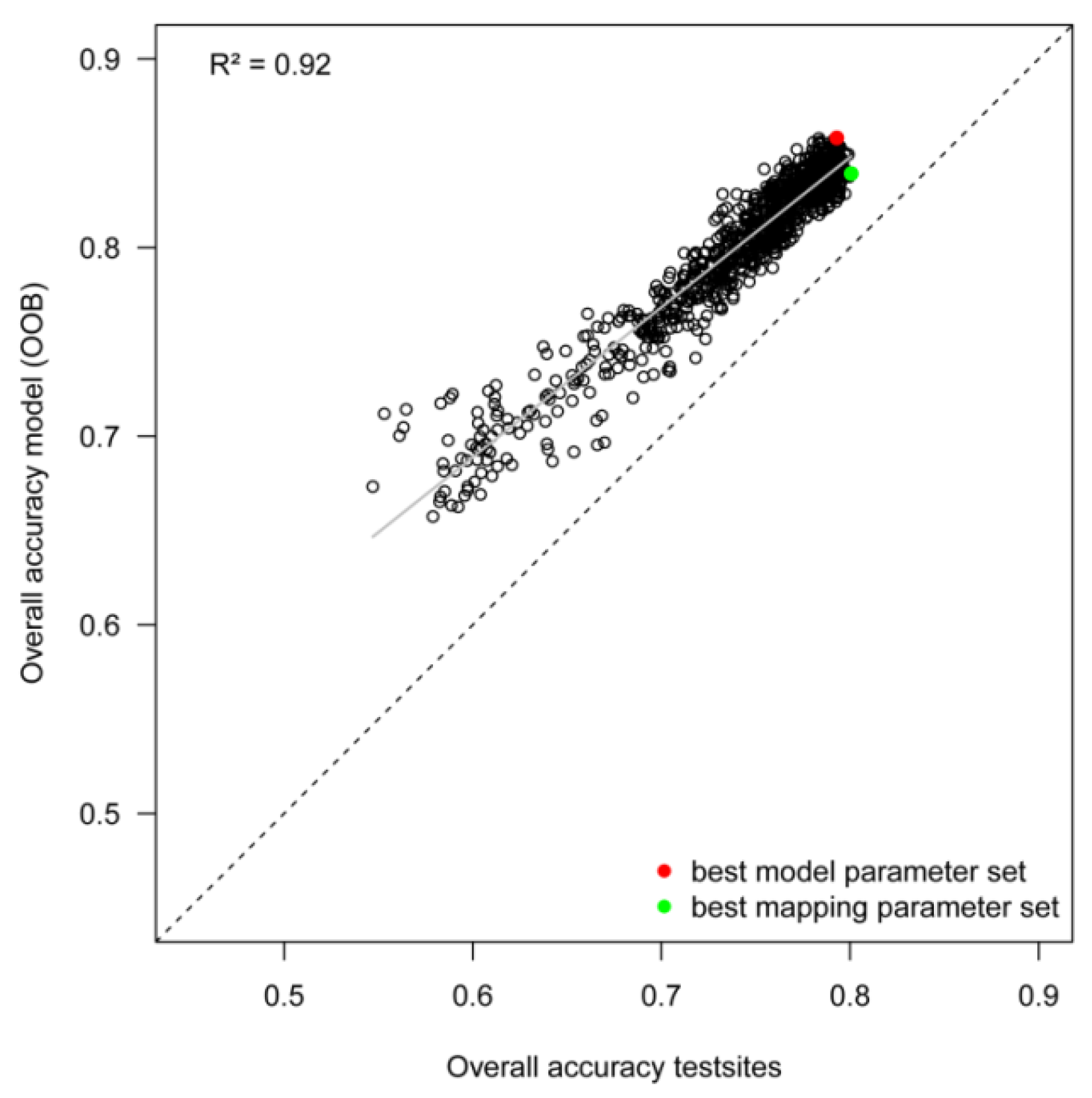
4.2. Model Robustness and Transferability to Test Sites
| Peanut | Sugarcane | Cassava | Soybean | Others | Sum | User’s Accuracy | |
|---|---|---|---|---|---|---|---|
| Peanut | 5.0 | 0.1 | 0.0 | 0.5 | 0.3 | 5.8 | 0.850 |
| Sugarcane | 1.7 | 179.6 | 0.6 | 6.9 | 78.9 | 267.6 | 0.671 |
| Cassava | 0.4 | 0.5 | 1.6 | 1.6 | 1.8 | 5.9 | 0.267 |
| Soybean | 2.5 | 6.5 | 7.3 | 174.3 | 41.1 | 231.6 | 0.752 |
| Others | 0.5 | 14.2 | 2.6 | 18.4 | 353.3 | 389.0 | 0.908 |
| Sum | 10.1 | 200.8 | 12.0 | 201.7 | 475.4 | 900.0 | |
| Producer’s accuracy | 0.491 | 0.894 | 0.132 | 0.864 | 0.743 | ||
| Overall accuracy (Kappa) | 0.793 (0.680) |
| Best Model Parameter Set | Best Mapping Parameter Set | ||
|---|---|---|---|
| scale | 70 | 65 | |
| shape | 0.50 | 0.70 | |
| compactness | 0.90 | 0.90 | |
| Overall accuracy | 0.793 | 0.801 | |
| Kappa | 0.680 | 0.692 | |
| Producer’s accuracy | Peanut | 0.491 | 0.637 |
| Sugarcane | 0.894 | 0.890 | |
| Cassava | 0.132 | 0.153 | |
| Soybean | 0.864 | 0.853 | |
| Others | 0.743 | 0.756 | |
| User’s accuracy | Peanut | 0.850 | 0.856 |
| Sugarcane | 0.671 | 0.673 | |
| Cassava | 0.267 | 0.281 | |
| Soybean | 0.752 | 0.782 | |
| Others | 0.908 | 0.907 | |
| Peanut | Sugarcane | Cassava | Soybean | Others | Sum | User’s Accuracy | |
|---|---|---|---|---|---|---|---|
| Peanut | 5.0 | 0.2 | 0.0 | 0.5 | 0.2 | 5.8 | 0.856 |
| Sugarcane | 1.5 | 180.0 | 0.5 | 8.9 | 76.7 | 267.6 | 0.673 |
| Cassava | 0.7 | 0.1 | 1.7 | 1.7 | 1.8 | 5.9 | 0.281 |
| Soybean | 0.4 | 8.1 | 6.8 | 181.2 | 35.1 | 231.6 | 0.782 |
| Others | 0.2 | 13.9 | 2.0 | 20.2 | 352.7 | 389.0 | 0.907 |
| Sum | 7.8 | 202.3 | 10.9 | 212.4 | 466.5 | 900.0 | |
| Producer’s accuracy | 0.637 | 0.890 | 0.153 | 0.853 | 0.756 | OA | 0.801 |
| Overall accuracy (Kappa) | 0.801 (0.692) |
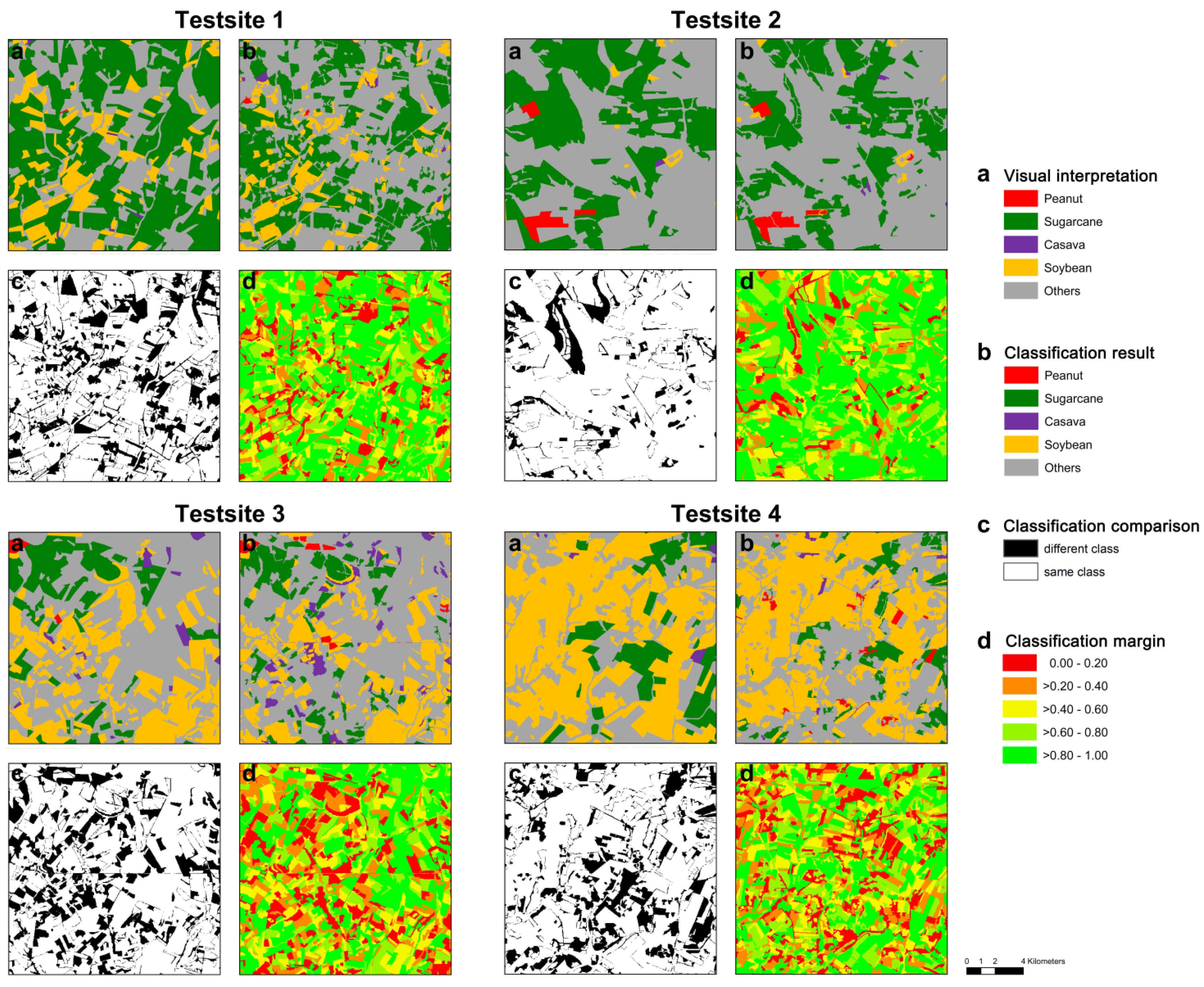
4.3. Exploitation of the Classification Margin

5. Conclusions
- Classification results depend to a high degree on the identification of suitable segmentation parameters. In our study, the worst segmentation yielded an overall accuracy (OA) of only 0.66 compared to the best result with OA 0.86 (with kappa of 0.48 and 0.79, respectively). This large spread in classification accuracies underlines that procedures are needed for (automatically) choosing suitable parameter settings. Today, this is mostly done in a manual way (trial-and-error), not necessarily leading to the best possible result.
- It is possible to fully automate the identification of suitable segmentation parameters (as well as input features for the classifier) using a full factorial design framework as chosen for this study (e.g., testing a pre-defined set of parameter combinations within fixed intervals each time optimizing input features using MDA information). The segmentation parameters derived from relatively few training samples were close to those one would have identified directly from another set of validation samples showing the robustness of the approach. The proposed optimization framework can be applied to all type of segmentation algorithms. Obviously, the simple feature selection approach chosen for our study can be replaced by other techniques. In our case, the resulting error surface (e.g., classification accuracy) was smooth enough to permit running a guided optimization procedure (Simplex or similar). It is not clear, however, if this would really speed-up the entire process.
- For regions similar to the Assis study area with sub-tropical climate, soybean and sugarcane can be classified with acceptable accuracy using two Landsat scenes acquired in January/February and August. However, using such acquisition dates, it was not possible to correctly map cassava and (to a lesser extend) peanut. The spectral signatures of these two crops were too similar for permitting a successful discrimination and mapping. Additional information would be required to map peanut and cassava with acceptable accuracy. The added-value of additional scenes also from other satellite sensors (e.g., Sentinel-2) deserves being tested in future work. In our study, additional scenes could not be processed due to persistent cloud coverage during the vegetative period.
- The classification margin—produced as a by-product using the Random Forest (RF) classifier—provides valuable information to the land cover (LC) map and deserves more attention. We were able to demonstrate (at pixel level) that the margin between first and second most often selected class (majority vote) closely reflects the accuracy of the final LC map. We found a close (linear) relation between the classification accuracy at pixel level and classification margin, with only 48% of the pixels being correctly classified in the worst case (margin <10%), while >92% of the pixels were correctly classified in the best case (margin >90%).
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Atzberger, C. Advances in remote sensing of agriculture: Context description, existing operational monitoring systems and major information needs. Remote Sens. 2013, 5, 949–981. [Google Scholar] [CrossRef]
- Becker-Reshef, I.; Justice, C.; Sullivan, M.; Vermote, E.; Tucker, C.; Anyamba, A.; Small, J.; Pak, E.; Masuoka, E.; Schmaltz, J.; et al. Monitoring global croplands with coarse resolution earth observations: The Global Agriculture Monitoring (GLAM) project. Remote Sens. 2010, 2, 1589–1609. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United Nations—Statistics Division. Available online: http://faostat3.fao.org/browse/Q/QC/E (accessed on 10 May 2015).
- Arvor, D.; Meirelles, M.; Dubreuil, V.; Bégué, A.; Shimabukuro, Y.E. Analyzing the agricultural transition in Mato Grosso, Brazil, using satellite-derived indices. Appl. Geogr. 2012, 32, 702–713. [Google Scholar] [CrossRef]
- Arvor, D.; Jonathan, M.; Meirelles, M.S.P.; Dubreuil, V.; Durieux, L. Classification of MODIS EVI time series for crop mapping in the state of Mato Grosso, Brazil. Int. J. Remote Sens. 2011, 32, 7847–7871. [Google Scholar] [CrossRef]
- Ippoliti-Ramilo, G.A.; Epiphanio, J.C.N.; Shimabukuro, Y.E. Landsat-5 Thematic Mapper data for pre-planting crop area evaluation in tropical countries. Int. J. Remote Sens. 2003, 24, 1521–1534. [Google Scholar] [CrossRef]
- Rizzi, R.; Rudorff, B.F.T. Estimativa da área de soja no Rio Grande do Sul por meio de imagens Landsat. Rev. Bras. Cartogr. 2005, 3, 226–234. [Google Scholar]
- Sugawara, L.M.; Rudorff, B.F.T.; Adami, M. Viabilidade de uso de imagens do Landsat em mapeamento de área cultivada com soja no Estado do Paraná. Pesqui. Agropecuária Bras. 2008, 43, 1763–1768. [Google Scholar] [CrossRef]
- Vieira, M.A.; Formaggio, A.R.; Rennó, C.D.; Atzberger, C.; Aguiar, D.A.; Mello, M.P. Object Based Image Analysis and Data Mining applied to a remotely sensed Landsat time-series to map sugarcane over large areas. Remote Sens. Environ. 2012, 123, 553–562. [Google Scholar] [CrossRef]
- Rudorff, B.F.T.; Aguiar, D.A.; Silva, W.F.; Sugawara, L.M.; Adami, M.; Moreira, M.A. Studies on the rapid expansion of sugarcane for ethanol production in São Paulo State (Brazil) using Landsat data. Remote Sens. 2010, 2, 1057–1076. [Google Scholar] [CrossRef]
- Justice, C.O.; Townshend, J.R.G.; Vermote, E.F.; Masuoka, E.; Wolfe, R.E.; Saleous, N.; Roy, D.P.; Morisette, J.T. An overview of MODIS Land data processing and product status. Remote Sens. Environ. 2002, 83, 3–15. [Google Scholar] [CrossRef]
- Justice, C.O.; Vermote, E.; Townshend, J.R.G.; DeFries, R.; Roy, D.P.; Hall, D.K.; Salomonson, V.V.; Privette, J.L.; Riggs, G.; Strahler, A.; et al. The Moderate Resolution Imaging Spectroradiometer (MODIS): Land remote sensing for global change research. IEEE Trans. Geosci. Remote Sens. 1998, 36, 1228–1249. [Google Scholar] [CrossRef]
- Brown, J.C.; Kastens, J.H.; Coutinho, A.C.; de Castro Victoria, D.; Bishop, C.R. Classifying multiyear agricultural land use data from Mato Grosso using time-series MODIS vegetation index data. Remote Sens. Environ. 2013, 130, 39–50. [Google Scholar] [CrossRef]
- Galford, G.L.; Mustard, J.F.; Melillo, J.; Gendrin, A.; Cerri, C.C.; Cerri, C.E.P. Wavelet analysis of MODIS time series to detect expansion and intensification of row-crop agriculture in Brazil. Remote Sens. Environ. 2008, 112, 576–587. [Google Scholar] [CrossRef]
- Rembold, F.; Atzberger, C.; Savin, I.; Rojas, O. Using low resolution satellite imagery for yield prediction and yield anomaly detection. Remote Sens. 2013, 5, 1704–1733. [Google Scholar] [CrossRef] [Green Version]
- Lamparelli, R.A.; de Carvalho, W.M.; Mercante, E. Mapeamento de semeaduras de soja (Glycine max (L.) Merr.) mediante dados MODIS/Terra e TM/Landsat 5: um comparativo. Eng. Agríc. 2008, 28, 334–344. [Google Scholar] [CrossRef]
- Rudorff, C.M.; Rizzi, R.; Rudorff, B.F.T.; Sugawara, L.M.; Vieira, C.A.O. Spectral-temporal response surface of MODIS sensor images for soybean area classification in Rio Grande do Sul State. Ciênc. Rural 2007, 37, 118–125. [Google Scholar] [CrossRef]
- Yi, J.L.; Shimabukuro, Y.E.; Quintanilha, J.A. Identificação e mapeamento de áreas de milho na região sul do Brasil utilizando imagens MODIS. Eng. Agríc. 2007, 27, 753–763. [Google Scholar] [CrossRef]
- Luiz, A.J.B.; Epiphanio, J.C.N. Amostragem por pontos em imagens de sensoriamento remoto para estimativa de área plantada por município. Simpósio Bras. Sensoriamento Remoto 2001, 10, 111–118. [Google Scholar]
- Atzberger, C.; Formaggio, A.R.; Shimabukuro, Y.E.; Udelhoven, T.; Mattiuzzi, M.; Sanchez, G.A.; Arai, E. Obtaining crop-specific time profiles of NDVI: the use of unmixing approaches for serving the continuity between SPOT-VGT and PROBA-V time series. Int. J. Remote Sens. 2014, 35, 2615–2638. [Google Scholar] [CrossRef]
- Lobell, D.B.; Asner, G.P. Cropland distributions from temporal unmixing of MODIS data. Remote Sens. Environ. 2004, 93, 412–422. [Google Scholar] [CrossRef]
- Ozdogan, M. The spatial distribution of crop types from MODIS data: Temporal unmixing using Independent Component Analysis. Remote Sens. Environ. 2010, 114, 1190–1204. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the blending of the Landsat and MODIS surface reflectance: Predicting daily Landsat surface reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar] [CrossRef]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Seitz, N.; White, J.C.; Gao, F.; Masek, J.G.; Stenhouse, G. Generation of dense time series synthetic Landsat data through data blending with MODIS using a spatial and temporal adaptive reflectance fusion model. Remote Sens. Environ. 2009, 113, 1988–1999. [Google Scholar] [CrossRef]
- Singh, D. Evaluation of long-term NDVI time series derived from Landsat data through blending with MODIS data. Atmósfera 2012, 25, 43–63. [Google Scholar]
- Wu, M.; Niu, Z.; Wang, C.; Wu, C.; Wang, L. Use of MODIS and Landsat time series data to generate high-resolution temporal synthetic Landsat data using a spatial and temporal reflectance fusion model. J. Appl. Remote Sens. 2012, 6, 063507–1. [Google Scholar]
- Cowen, D.J.; Jensen, J.R.; Bresnahan, P.J.; Ehler, G.B.; Graves, D.; Huang, X.; Wiesner, C.; Mackey, H.E. The design and implementation of an integrated geographic information system for environmental applications. Photogramm. Eng. Remote Sens. 1995, 61, 1393–1404. [Google Scholar]
- Powell, S.L.; Pflugmacher, D.; Kirschbaum, A.A.; Kim, Y.; Cohen, W.B. Moderate resolution remote sensing alternatives: A review of Landsat-like sensors and their applications. J. Appl. Remote Sens. 2007. [Google Scholar] [CrossRef]
- Whitcraft, A.K.; Becker-Reshef, I.; Killough, B.D.; Justice, C.O. Meeting earth observation requirements for global agricultural monitoring: An evaluation of the revisit capabilities of current and planned moderate resolution optical earth observing missions. Remote Sens. 2015, 7, 1482–1503. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Hay, G.J.; Castilla, G. Geographic Object-Based Image Analysis (GEOBIA): A new name for a new discipline. In Object-Based Image Analysis; Blaschke, T., Lang, S., Hay, G.J., Eds.; Lecture Notes in Geoinformation and Cartography; Springer: Berlin/Heidelberg, Germany, 2008; pp. 75–89. [Google Scholar]
- Löw, F.; Duveiller, G. Defining the spatial resolution requirements for crop identification using optical remote sensing. Remote Sens. 2014, 6, 9034–9063. [Google Scholar] [CrossRef]
- Myint, S.W.; Gober, P.; Brazel, A.; Grossman-Clarke, S.; Weng, Q. Per-pixel vs. object-based classification of urban land cover extraction using high spatial resolution imagery. Remote Sens. Environ. 2011, 115, 1145–1161. [Google Scholar] [CrossRef]
- Toscani, P.; Immitzer, M.; Atzberger, C. Texturanalyse mittels diskreter Wavelet Transformation für die objektbasierte Klassifikation von Orthophotos. Photogramm. Fernerkund. Geoinf. 2013, 2, 105–121. [Google Scholar] [CrossRef]
- Devadas, R.; Denham, R.J.; Pringle, M. Support vector machine classification of object-based data for crop mapping, using multi-temporal Landsat imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 39, 185–190. [Google Scholar] [CrossRef]
- Dronova, I.; Gong, P.; Clinton, N.E.; Wang, L.; Fu, W.; Qi, S.; Liu, Y. Landscape analysis of wetland plant functional types: The effects of image segmentation scale, vegetation classes and classification methods. Remote Sens. Environ. 2012, 127, 357–369. [Google Scholar] [CrossRef]
- Formaggio, A.R.; Vieira, M.A.; Renno, C.D. Object Based Image Analysis (OBIA) and Data Mining (DM) in Landsat time series for mapping soybean in intensive agricultural regions. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Munich, Germany, 22–27 July 2012; pp. 2257–2260.
- Formaggio, A.R.; Vieira, M.A.; Rennó, C.D.; Aguiar, D.A.; Mello, M.P. Object-Based Image Analysis and Data Mining for mapping sugarcane with Landsat imagery in Brazil. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2010, 38, 553–562. [Google Scholar]
- Long, J.A.; Lawrence, R.L.; Greenwood, M.C.; Marshall, L.; Miller, P.R. Object-oriented crop classification using multitemporal ETM+ SLC-off imagery and random forest. GIScience Remote Sens. 2013, 50, 418–436. [Google Scholar]
- Peña-Barragán, J.M.; Ngugi, M.K.; Plant, R.E.; Six, J. Object-based crop identification using multiple vegetation indices, textural features and crop phenology. Remote Sens. Environ. 2011, 115, 1301–1316. [Google Scholar] [CrossRef]
- Peña, J.M.; Gutiérrez, P.A.; Hervás-Martínez, C.; Six, J.; Plant, R.E.; López-Granados, F. Object-based image classification of summer crops with machine learning methods. Remote Sens. 2014, 6, 5019–5041. [Google Scholar] [CrossRef]
- Dey, V.; Zhang, Y.; Zhong, M. A review on image segmentation techniques with remote sensing perspective. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2010, 38, 31–42. [Google Scholar]
- Forster, D.; Kellenberger, T.W.; Buehler, Y.; Lennartz, B. Mapping diversified peri-urban agriculture—Potential of object-based versus per-field land cover/land use classification. Geocarto Int. 2010, 25, 171–186. [Google Scholar] [CrossRef]
- Muñoz, X.; Freixenet, J.; Cufı́, X.; Martı́, J. Strategies for image segmentation combining region and boundary information. Pattern Recognit. Lett. 2003, 24, 375–392. [Google Scholar] [CrossRef]
- Yan, L.; Roy, D.P. Automated crop field extraction from multi-temporal Web Enabled Landsat Data. Remote Sens. Environ. 2014, 144, 42–64. [Google Scholar] [CrossRef]
- Baatz, M.; Schäpe, A. Multiresolution Segmentation: An optimization approach for high quality multi-scale image segmentation. In Angewandte Geographische Informationsverarbeitung XII; Strobl, J., Blaschke, T., Griesebner, G., Eds.; Wichmann Verlag: Karlsruhe, Germany, 2000; pp. 12–23. [Google Scholar]
- Liu, D.; Xia, F. Assessing object-based classification: Advantages and limitations. Remote Sens. Lett. 2010, 1, 187–194. [Google Scholar] [CrossRef]
- Stumpf, A.; Kerle, N. Object-oriented mapping of landslides using Random Forests. Remote Sens. Environ. 2011, 115, 2564–2577. [Google Scholar] [CrossRef]
- Taşdemir, K.; Milenov, P.; Tapsall, B. A hybrid method combining SOM-based clustering and object-based analysis for identifying land in good agricultural condition. Comput. Electron. Agric. 2012, 83, 92–101. [Google Scholar] [CrossRef]
- Benz, U.C.; Hofmann, P.; Willhauck, G.; Lingenfelder, I.; Heynen, M. Multi-resolution, object-oriented fuzzy analysis of remote sensing data for GIS-ready information. ISPRS J. Photogramm. Remote Sens. 2004, 58, 239–258. [Google Scholar] [CrossRef]
- Bo, S.; Ding, L.; Li, H.; Di, F.; Zhu, C. Mean shift-based clustering analysis of multispectral remote sensing imagery. Int. J. Remote Sens. 2009, 30, 817–827. [Google Scholar] [CrossRef]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Beucher, S. The watershed transformation applied to image segmentation. Scanning Microsc. Suppl. 1992, 6, 299–299. [Google Scholar]
- Vantaram, S.R.; Saber, E. Survey of contemporary trends in color image segmentation. J. Electron. Imaging 2012. [Google Scholar] [CrossRef]
- Oliveira, J.C.; Formaggio, A.R.; Epiphanio, J.C.N.; Luiz, A.J.B. Index for the Evaluation of Segmentation (IAVAS): An application to agriculture. Mapp. Sci. Remote Sens. 2003, 40, 155–169. [Google Scholar] [CrossRef]
- Stefanski, J.; Mack, B.; Waske, B. Optimization of object-based image analysis with random forests for land cover mapping. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 2492–2504. [Google Scholar] [CrossRef]
- IBGE Sistema IBGE de Recuperação Automática. Available online: http://www.sidra.ibge.gov.br (accessed on 8 March 2015).
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; de Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- Aguiar, D.A.; Rudorff, B.F.T.; Silva, W.F.; Adami, M.; Mello, M.P. Remote sensing images in support of environmental protocol: Monitoring the sugarcane harvest in São Paulo State, Brazil. Remote Sens. 2011, 3, 2682–2703. [Google Scholar] [CrossRef]
- De Aguiar, D.A.; Rudorff, B.T.F.; Rizzi, R.; Shimabukuro, Y.E. Monitoramento da colheita da cana-de-açúcar por meio de imagens MODIS. Rev. Bras. Cartogr. 2008, 4, 375–383. [Google Scholar]
- IEA Intituto de economia agricola: Área e Produção de milho de primeira safra, de segunda safra e irrigado, em 2014, no estado de São Paulo. Available online: http://ciagri.iea.sp.gov.br/nia1/ (accessed on 8 March 2015).
- Roy, D.P.; Wulder, M.A.; Loveland, T.R.; Woodcock, C.E.; Allen, R.G.; Anderson, M.C.; Helder, D.; Irons, J.R.; Johnson, D.M.; Kennedy, R.; et al. Landsat-8: Science and product vision for terrestrial global change research. Remote Sens. Environ. 2014, 145, 154–172. [Google Scholar] [CrossRef]
- Cihlar, J. Land cover mapping of large areas from satellites: Status and research priorities. Int. J. Remote Sens. 2000, 21, 1093–1114. [Google Scholar] [CrossRef]
- INPE. Available online: http://cbers2.dpi.inpe.br/ (accessed on 8 March 2015).
- Sanches, I.D.; Epiphanio, J.C.N.; Formaggio, A.R. Culturas agrícolas em imagens multitemporais do satélite Landsat. Agric. Em São Paulo 2005, 52, 83–96. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with Random Forest using very high spatial resolution 8-band WorldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Rodríguez-Galiano, V.F.; Chica-Olmo, M.; Abarca-Hernandez, F.; Atkinson, P.M.; Jeganathan, C. Random Forest classification of Mediterranean land cover using multi-seasonal imagery and multi-seasonal texture. Remote Sens. Environ. 2012, 121, 93–107. [Google Scholar] [CrossRef]
- Waske, B.; Braun, M. Classifier ensembles for land cover mapping using multitemporal SAR imagery. ISPRS J. Photogramm. Remote Sens. 2009, 64, 450–457. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C. Early Detection of Bark Beetle Infestation in Norway Spruce (Picea abies, L.) using WorldView-2 Data. Photogramm. Fernerkund. Geoinf. 2014, 2014, 351–367. [Google Scholar] [CrossRef]
- Breiman, L. Manual on Setting up, Using, and Understanding Random Forests V3.1; University of California at Berkeley: Berkeley, CA, USA, 2002. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Vuolo, F.; Atzberger, C. Improving land cover maps in areas of disagreement of existing products using NDVI time series of MODIS—Example for Europe. Photogramm. Fernerkund. Geoinf. 2014, 2014, 393–407. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling R package. 2014.
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z. Caret: Classification and Regression Training R package. 2014.
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schultz, B.; Immitzer, M.; Formaggio, A.R.; Sanches, I.D.A.; Luiz, A.J.B.; Atzberger, C. Self-Guided Segmentation and Classification of Multi-Temporal Landsat 8 Images for Crop Type Mapping in Southeastern Brazil. Remote Sens. 2015, 7, 14482-14508. https://doi.org/10.3390/rs71114482
Schultz B, Immitzer M, Formaggio AR, Sanches IDA, Luiz AJB, Atzberger C. Self-Guided Segmentation and Classification of Multi-Temporal Landsat 8 Images for Crop Type Mapping in Southeastern Brazil. Remote Sensing. 2015; 7(11):14482-14508. https://doi.org/10.3390/rs71114482
Chicago/Turabian StyleSchultz, Bruno, Markus Immitzer, Antonio Roberto Formaggio, Ieda Del’ Arco Sanches, Alfredo José Barreto Luiz, and Clement Atzberger. 2015. "Self-Guided Segmentation and Classification of Multi-Temporal Landsat 8 Images for Crop Type Mapping in Southeastern Brazil" Remote Sensing 7, no. 11: 14482-14508. https://doi.org/10.3390/rs71114482








