1. Introduction
The reproducibility and replication (R&R) of scientific findings has recently moved to the forefront of research agenda in many fields [
1,
2,
3,
4,
5] since it has been discovered that findings often cannot be reproduced or replicated [
5,
6]. While the two “R’s”—reproducibility and replicability—are intertwined, there are key differences between their goals. Adopting the definitions from the National Science Foundation [
7] and the National Academy of Science, Engineering, and Medicine [
8], we define reproducibility as the ability of a researcher to duplicate the results of a prior study using the same data and methods as the original investigator. In short, if a researcher makes the data and methods/code available, another researcher should be able to produce the exact same results. In comparison, replicability is the ability of a researcher to duplicate results using similar methods but with new data.
Achieving R&R is critical for advancing scientific discoveries, yet neither topic has received much attention in geography and the spatial sciences, where investigations tend to be observational instead of experimental or theoretical [
9]. R&R has received even less attention in remote sensing (but see [
10] for an early take), even though the field is uniquely positioned to contribute to R&R on several fronts. First, there is a rich archive of publicly available remote sensing datasets (e.g., Landsat), supporting opportunities for reproducibility [
9,
11]. Second, remote sensing studies, and in particular hyperspectral studies, are often situated in extremely local contexts (e.g., agricultural plots) due to the need for ground reference data and the high labor and time costs of operating equipment. Yet, an implicit goal of science is to develop widely applicable methodologies and generalizable findings that can be applied in different contexts. Thus, working toward the replicability of methods and findings across different study areas is important for advancing remote sensing science.
Despite the myriad opportunities for remote sensing scientists to explore R&R issues, very few formal efforts have been documented. One reason is likely because remote sensing scientists often work with large datasets and perform complex spectral and spatial manipulations [
12,
13,
14,
15,
16], which makes R&R difficult if processing code is not made available. Until recently, many scientific publications did not require code to be submitted as part of the manuscript review process, although this is changing. Replication in remote sensing is also hindered by attributes of local environments, which makes the transfer of results from one landscape to another difficult. However, if we are to develop methodologies that are transferrable across space, it is necessary to begin developing and implementing protocols for testing the R&R of remote sensing studies. One way to do this is to incorporate multi-field, multi-environment analyses into studies to self-test the replicability of methods and results.
Precision agriculture is one field where immediate gains can be made toward testing the replicability of methods while also contributing a larger understanding of the extent of R&R issues in remote sensing. Since the overall goal of precision agriculture is to decrease the ambiguity of decisions required on agricultural lands that are often highly variable [
17], the ability to transfer methods and findings from one environment or location to another requires them to be replicable [
18]. However, most studies capture data in a single region or location (often in a single crop field) under uniform conditions [
12,
15], thus limiting their generalizability across environmental or geographical contexts. Furthermore, the implicit assumption is that methods and findings are extendable beyond the single field in which they were tested, but in most cases, no such evidence is provided. Many studies lack basic explanation for environmental variances such as soil, hydrology, and topography that can cause reflectance variations, thereby altering results across space [
16]. Ultimately, remote sensing methodologies are of little practical value for precision agriculture if they are developed, tested, and applicable in a single location where these multiple and often confounding factors are held constant.
Partial least squares (PLS) regression has become an accepted technique in vegetation studies using hyperspectral data for estimating a range of biophysical and biochemical properties [
19,
20,
21,
22,
23]. In situations where the number of independent variables is large and the variables are collinear, which is common with hyperspectral data, multiple linear regression will often overfit the model [
24,
25]. PLS regression standardizes model construction from the preprocessed hyperspectral data via latent variables, from which the predictive capabilities of the model can be tested. Recently, variations of PLS regression using a waveband selection procedure [
13] have been proposed and adopted, but there has been little effort to test the replicability of these methods across environments to determine whether results might be transferable.
The objective of this study is to investigate the replicability of PLS regression methods, including PLS with waveband selection, for predicting nutrient content in plant and grain material across multiple environments. This study addresses gaps in the remote sensing R&R literature by replicating a methodological workflow using hyperspectral data and PLS regression for predicting nutrients in a single crop but in two varying environments in different international contexts to determine the degree to which the methods are replicable. The focus is on
Eragrostis tef (tef), a cereal crop primarily grown in Ethiopia, although production has been expanding to other parts of the world due to its versatility and resistance to drought. Tef is ideal for studying replicability because it is grown in different international contexts and is known for being successfully cultivated across differing environments. Additionally, very few hyperspectral analyses have been performed on non-milled grains [
26], so this study contributes knowledge in that realm as well.
Tef is a grass (Family: Poaceae) that has received little attention from the remote sensing and precision agricultural communities despite its versatile cultivation characteristics. Tef is thought to be one of the earliest domesticated plants [
27], with the center of origin and diversity in Ethiopia/Horn of Africa [
28]. It is drought and heat resistant, has a high nutrient content, and is grown for animal feed as well as a staple food crop [
29]. While tef can be cultivated across many environments, it is primarily grown in Ethiopia, where it is the most commonly harvested crop, popular for its highly nutritious, gluten-free grain [
30,
31,
32,
33,
34,
35]. Recently, cultivation has been spreading outside the region; in the United States, tef is planted as a sequential forage crop for livestock feed but is currently only grown in a handful of locations [
29,
36].
3. Analytical Methods
Partial least squares (PLS) regression was implemented to assess the relationship between reflectance (independent variable) and nutrient content (dependent variable) of both the plant and grain. PLS, which can also stand for projection to latent structures [
43], was selected over other forms of regression because it accounts for overfitting errors common when analyzing hyperspectral data [
13,
44]. Briefly, PLS regression finds a set of components (called latent factors) from
X, a matrix of predictors collected on the observations, that best predict
Y, a matrix of dependent observations [
43]. These latent factors, or latent vectors, are orthogonal, and thus explain as much of the covariance between
X and
Y as possible, often resulting in a smaller number of variables than principal component regression. PLS regression extracts
X-scores from the latent variables to construct a model to predict the
Y-scores. In PLS, the
X- and
Y-scores are subject to redundancy analysis that seeks directionality in factor space until the most accurate prediction is found [
25,
45]. When implementing PLS regression with hyperspectral data, it is important to ensure the number of latent variables does not far exceed the number of independent variables being used, as overfitting can occur [
13].
3.1. Specification of the PLS Regression Model Using the Full Spectrum
The PLS equation follows a standard regression (Equation (1)):
where the response variable
is the nutrient value, and the predictor variables
to
are the reflectance (SG) or derivative (FD) values for bands 1 to
(here, 277).
to
are the estimated weighted regression coefficients computed directly from the PLS loadings corresponding to the model with the optimal number of latent variables, and
is the error vector. The optimal number of latent variables (NLV) is determined through a leave-one-out (LOO) cross-validation and assessed through the minimum root mean square error:
where
represents the predicted response when the model is built without sample
i,
represents the measured nutrient value for sample
i, and
n represents the number of samples used in the calibration [
46]. Twelve, standard PLS regressions were performed, each corresponding to a dataset in
Table 1, and each including the full set of 277 bands (PLS-Full).
3.2. PLS Regression with Waveband Selection
A modified form of PLS regression, known as the waveband selection (or iterative stepwise elimination) method, was developed to eliminate noisy and unhelpful predictors in hyperspectral studies [
13,
44]. Instead of including all 277 wavebands, the number is reduced iteratively [
44] by dropping the least important wavebands (similar to stepwise linear regression). Waveband importance (
) is determined as:
where
and
are the regression coefficient and standard deviation corresponding to waveband
k. The selection begins with all 277 wavebands, and the waveband contributing least to the model (lowest
) is removed. The model (Equation (1)) is then re-run with 276 variables, and so on, until the maximum predictive capability is achieved [
47]. A representation of the iterative processes to determine the maximum predictive capability is shown in
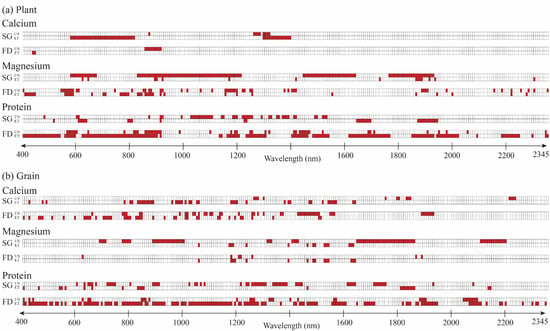
Figure 5. This version of the model is hereafter referred to as PLS-Wave.
3.3. Predictive Ability of PLS Regression
To test the predictive capabilities of the PLS-Full and PLS-Wave models, we implemented a bootstrapping procedure by dividing the data into calibration (65–75%) and validation (25–35%) sets replacing the data of these sets
times (following [
48,
49]). After each separation, the models were calibrated (assessed through
RMSECV) and then validated using root mean square error of prediction (
RMSEP):
where
represents the predicted nutrient values,
represents the measured nutrient values, and
n represents the number of samples in the validation subset [
46]. Mean coefficient of determination (
R2),
R2 standard deviation (
R2 std.), and
RMSEP standard deviation (
RMSEP std) are also reported for the validation. PLS regressions were performed in Matlab v2016a (MathWorks, Sherborn, MA, USA).
3.4. Replication across Environments
To assess the replicability of the PLS-Full and PLS-Wave regression methodologies for predicting nutrient content across environments, we compared model performance between the US and ET using a difference of means (
t-test) for each component–nutrient combination (e.g., grain–calcium, plant–calcium, etc.) between the two sites from the bootstrapped results. To compare the similarity of wavebands selected by the PLS-Wave model, the Jaccard index [
50] was used to measure the overlap in selected wavebands compared to the total number of wavebands selected for each site (Equation (5)):
where
A and
B are the set of selected wavebands in the two locations, respectively. A Jaccard value of 1.0 indicates that the models for the two locations overlap completely in terms of the wavebands selected as important for prediction; 0 indicates the two locations share none of the same wavebands.
6. Conclusions
This study investigated the replicability of PLS regression methods, including PLS with waveband selection, for predicting nutrient content in plant and grain material across multiple environments. Using Eragrostis tef (tef) as a target crop, this study compared PLS model fits and selected wavebands across two environments in the U.S. and Ethiopia to determine the extent to which the methods and finding are replicable. Three main findings emerge from this study:
First, the model fits and wavebands selected as important for nutrient prediction were not replicable across the two study sites in the US and Ethiopia. Eleven of the 12 comparisons had statistically different model fits across 1000 bootstrapped iterations, and the Jaccard index for similarity indicated very low similarities in the wavebands selected.
Combining samples from both environments improved model fits, suggesting that increasing within-sample variation may improve the predictability of PLS models across study areas, though caution is reserved if great disparities in sample values are great. Our recommendation is to build a more open and transparent culture of data sharing within the remote sensing community that will permit data sharing in order to advance modeling capabilities and promote development of more generalizable predictive models.
Results using PLS regression with hyperspectral data from non-milled grains were generally positive, and wavebands for protein prediction generally agreed with other studies. While more research is needed to determine whether these consistencies are true positives or are affected by other factors, this study contributes to the gap in the literature related to non-milled grains.