Assimilating Remote Sensing Phenological Information into the WOFOST Model for Rice Growth Simulation
Abstract
1. Introduction
2. Study Area and Data
2.1. Study Area
2.2. Field Data
2.3. Remote Sensing Data
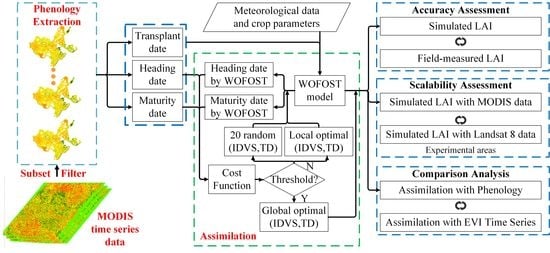
3. Methods
3.1. Phenology Development Simulation in the WOFOST Model
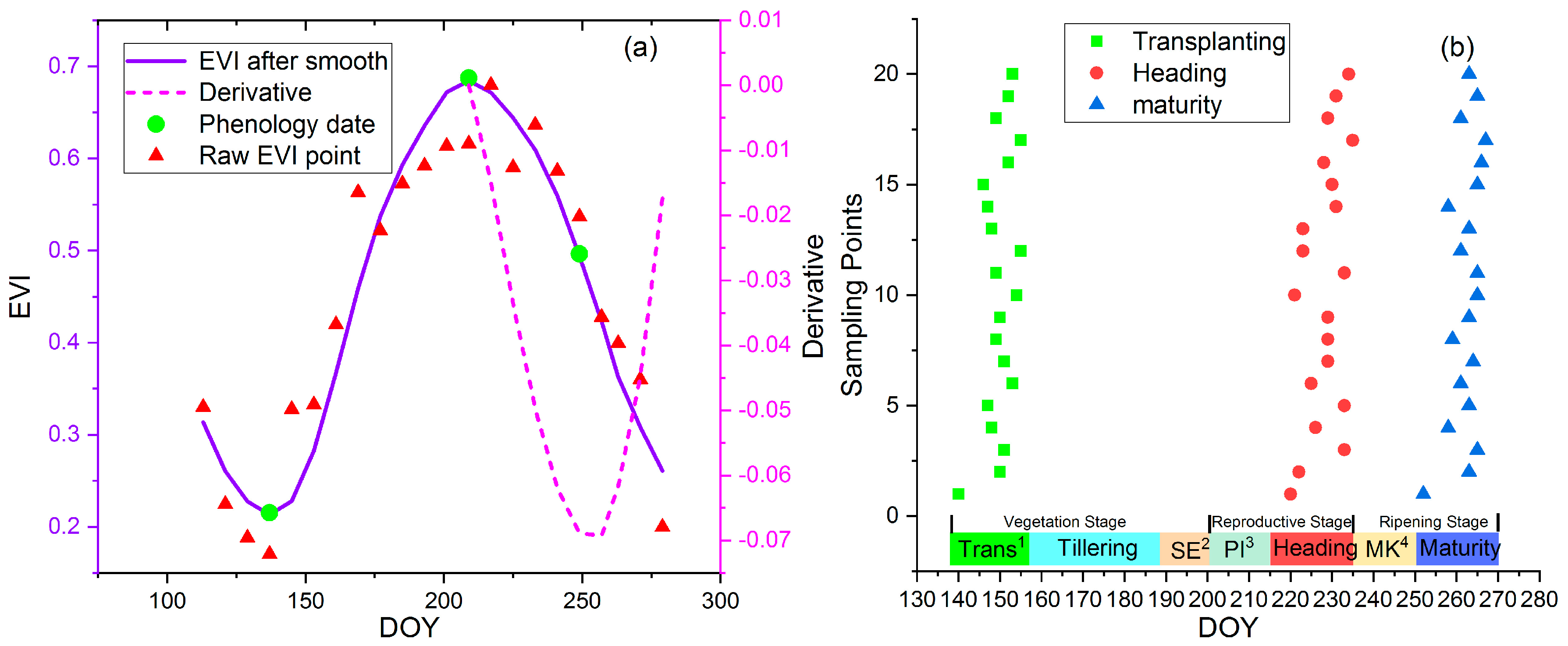
3.2. Phenology Extraction from Remote Sensing Data
3.3. Assimilation of Remote Sensing Phenological Information with the WOFOST Model
4. Results
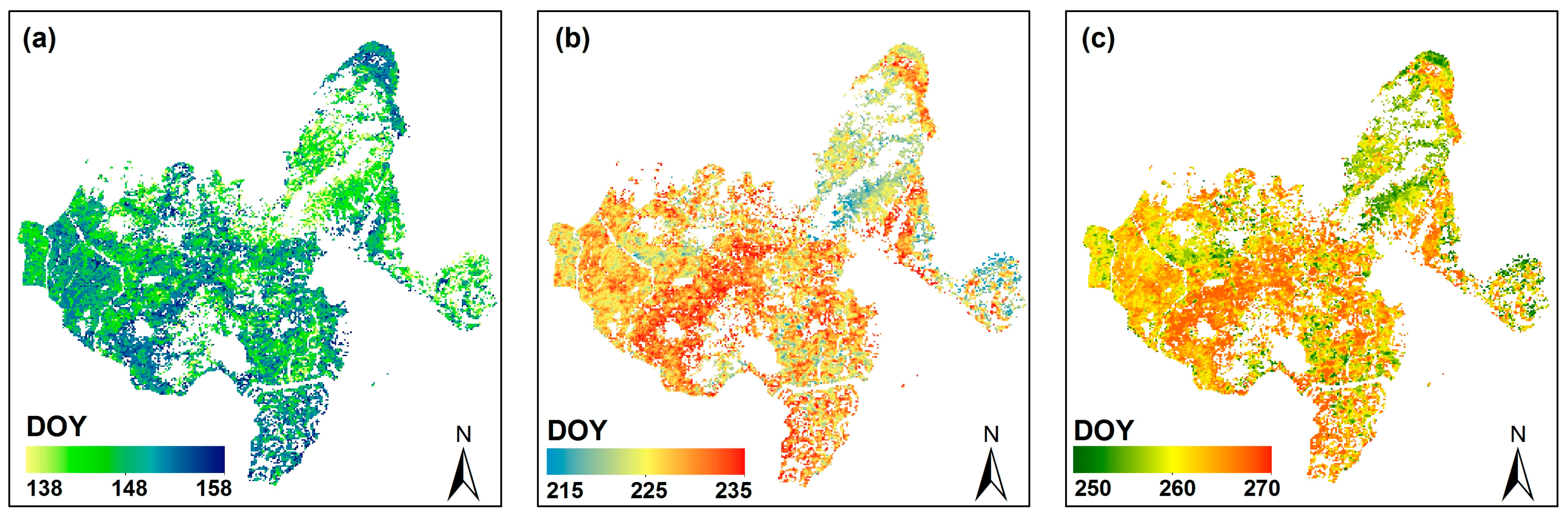
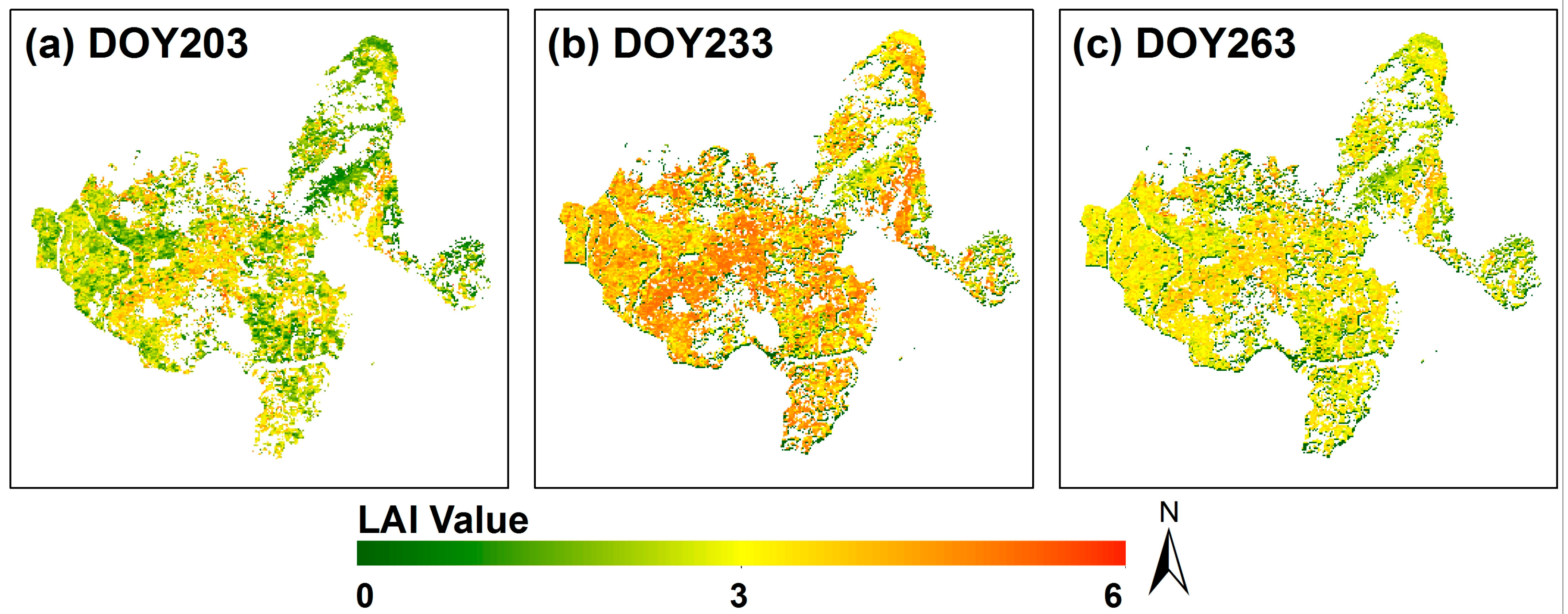
4.1. Remote Sensing Phenology Extraction
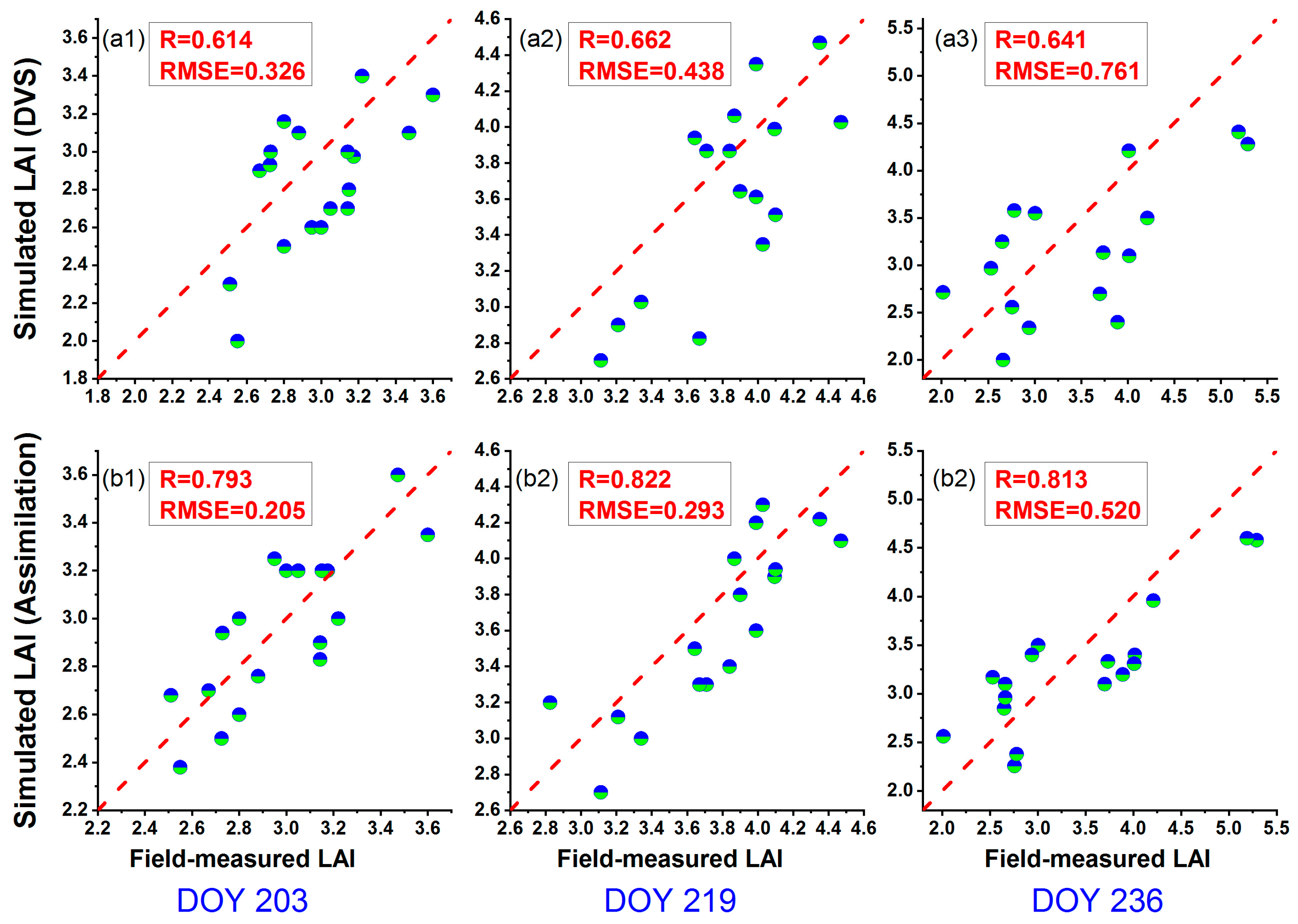
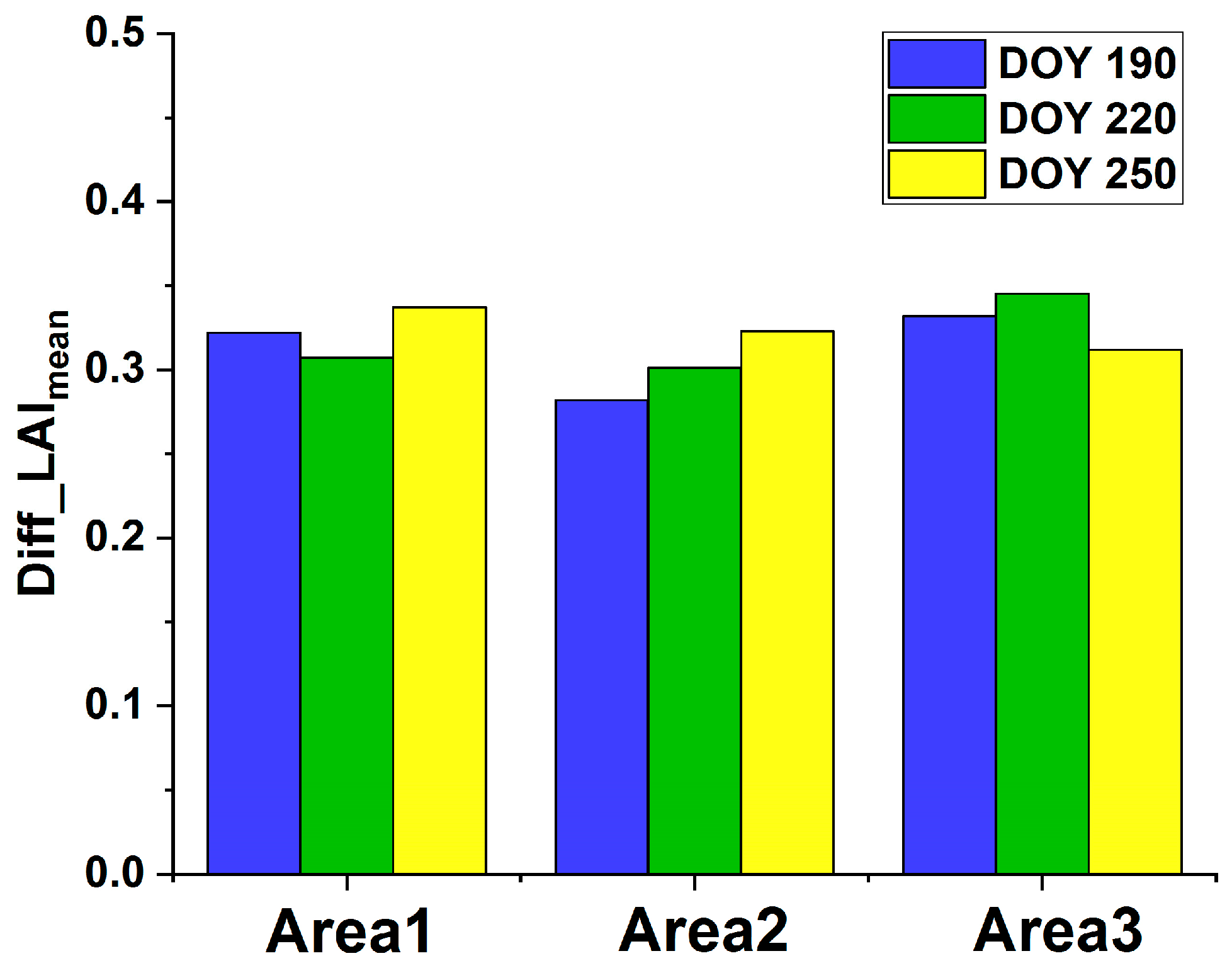
4.2. Rice Growth Simulation with Assimilated Phenological Information
4.3. Comparison with EVI-Assimilated Rice Growth Simulation
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, J.S.; Huang, J.X.; Hui, L.; Pei, Z. Rice yield estimation by assimilation remote sensing into crop growth modell. Sci. Sin. 2010, 40, 177–187. [Google Scholar]
- Huang, J.; Tian, L.; Liang, S.; Ma, H.; Becker-Reshef, I.; Huang, Y.; Su, W.; Zhang, X.; Zhu, D.; Wu, W. Improving winter wheat yield estimation by assimilation of the leaf area index from Landsat TM and MODIS data into the WOFOST model. Agric. For. Meteorol. 2015, 204, 106–121. [Google Scholar] [CrossRef]
- Mishra, S.K.; Shekh, A.M.; Pandey, V.; Yadav, S.B.; Patel, H.R. Sensitivity analysis of four wheat cultivars to varying photoperiod and temperature at different phenological stages using WOFOST model. J. Agrometeorol. 2015, 17, 74–79. [Google Scholar]
- Heulen, H.V.; Van Diepen, C. Crop growth models and agro-ecological characterization. In Proceedings of the First Congress of the European Society of Agronomy, Paris, France, 5–7 December 1990; pp. 2–16. [Google Scholar]
- Bouman, B.A.M.; Keulen, H.V.; Laar, H.H.V.; Rabbinge, R. The ‘School of de Wit’ crop growth simulation models: A pedigree and historical overview. Agric. Syst. 1996, 52, 171–198. [Google Scholar] [CrossRef]
- Jin, X.; Kumar, L.; Li, Z.; Feng, H.; Xu, X.; Yang, G.; Wang, J. A review of data assimilation of remote sensing and crop models. Eur. J. Agron. 2018, 92, 141–152. [Google Scholar] [CrossRef]
- Wu, L.; Liu, X.; Wang, P.; Zhou, B.; Liu, M.; Li, X. The assimilation of spectral sensing and the WOFOST model for the dynamic simulation of cadmium accumulation in rice tissues. Int. J. Appl. Earth Observ. Geoinform. 2013, 25, 66–75. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, S.; Shen, S. Assimilating remote sensing information with crop model using Ensemble Kalman Filter for improving LAI monitoring and yield estimation. Ecol. Model. 2013, 270, 30–42. [Google Scholar] [CrossRef]
- Mladen, T.; Rossella, A.; Ljubomir, Z.; Mariethereseabi, S.; Claudio, S.; Pasquale, S. Assessment of AquaCrop, CropSyst, and WOFOST Models in the Simulation of Sunflower Growth under Different Water Regimes. Agron. J. 2014, 101, 509–521. [Google Scholar]
- Dorigo, W.A.; Zurita-Milla, R.; Wit, A.J.W.D.; Brazile, J.; Singh, R.; Schaepman, M.E. A review on reflective remote sensing and data assimilation techniques for enhanced agroecosystem modeling. Int. J. Appl. Earth Observ. Geoinform. 2007, 9, 165–193. [Google Scholar] [CrossRef]
- Liang, S.; Qin, J. Data Assimilation Methods for Land Surface Variable Estimation; Springer: Amsterdam, The Netherlands, 2008; pp. 313–339. [Google Scholar]
- Ahmadalipour, A.; Moradkhani, H.; Yan, H.; Zarekarizi, M. Remote sensing of drought: Vegetation, soil moisture, and data assimilation. In Remote Sensing of Hydrological Extremes; Springer: Berlin, Germany, 2017; pp. 121–149. [Google Scholar]
- Chen, Y.; Zhang, Z.; Tao, F. Improving regional winter wheat yield estimation through assimilation of phenology and leaf area index from remote sensing data. Eur. J. Agron. 2018, 101, 163–173. [Google Scholar] [CrossRef]
- Maas, S.J. Use of remotely-sensed information in agricultural crop growth models. Ecol. Model. 1988, 41, 247–268. [Google Scholar] [CrossRef]
- Moulin, S.; Bondeau, A.; Delecolle, R. Combining agricultural crop models and satellite observations: From field to regional scales. Int. J. Remote Sens. 1998, 19, 1021–1036. [Google Scholar] [CrossRef]
- Huang, J.; Ma, H.; Su, W.; Zhang, X.; Huang, Y.; Fan, J.; Wu, W. Jointly assimilating MODIS LAI and ET products into the SWAP model for winter wheat yield estimation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4060–4071. [Google Scholar] [CrossRef]
- Huang, J.; Sedano, F.; Huang, Y.; Ma, H.; Li, X.; Liang, S.; Tian, L.; Zhang, X.; Fan, J.; Wu, W. Assimilating a synthetic Kalman filter leaf area index series into the WOFOST model to improve regional winter wheat yield estimation. Agric. For. Meteorol. 2016, 216, 188–202. [Google Scholar] [CrossRef]
- Huang, J.; Ma, H.; Sedano, F.; Lewis, P.; Liang, S.; Wu, Q.; Su, W.; Zhang, X.; Zhu, D. Evaluation of regional estimates of winter wheat yield by assimilating three remotely sensed reflectance datasets into the coupled WOFOST–PROSAIL model. Eur. J. Agron. 2019, 102, 1–13. [Google Scholar] [CrossRef]
- Bouman, B.A.M. Linking physical remote sensing models with crop growth simulation models, applied for sugar beet. Int. J. Remote Sens. 1992, 13, 2565–2581. [Google Scholar] [CrossRef]
- Weiss, M.; Troufleau, D.; Baret, F.; Chauki, H.; Prévot, L.; Olioso, A.; Bruguier, N.; Brisson, N. Coupling canopy functioning and radiative transfer models for remote sensing data assimilation. Agric. For. Meteorol. 2001, 108, 113–128. [Google Scholar] [CrossRef]
- Li, H.; Chen, Z.; Liu, G.; Jiang, Z.; Huang, C. Improving Winter Wheat Yield Estimation from the CERES-Wheat Model to Assimilate Leaf Area Index with Different Assimilation Methods and Spatio-Temporal Scales. Remote Sens. 2017, 9, 190. [Google Scholar] [CrossRef]
- Jin, M.; Liu, X.; Wu, L.; Liu, M. An improved assimilation method with stress factors incorporated in the WOFOST model for the efficient assessment of heavy metal stress levels in rice. Int. J. Appl. Earth Observ. Geoinform. 2015, 41, 118–129. [Google Scholar] [CrossRef]
- Cheng, Z.; Meng, J.; Wang, Y. Improving Spring Maize Yield Estimation at Field Scale by Assimilating Time-Series HJ-1 CCD Data into the WOFOST Model Using a New Method with Fast Algorithms. Remote Sens. 2016, 8, 303. [Google Scholar] [CrossRef]
- Huang, X.; Huang, G.; Yu, C.; Ni, S.; Yu, L. A multiple crop model ensemble for improving broad-scale yield prediction using Bayesian model averaging. Field Crops Res. 2017, 211, 114–124. [Google Scholar] [CrossRef]
- Liu, F.; Liu, X.; Ding, C.; Wu, L. The dynamic simulation of rice growth parameters under cadmium stress with the assimilation of multi-period spectral indices and crop model. Field Crops Res. 2015, 183, 225–234. [Google Scholar] [CrossRef]
- Wu, L.; Liu, X.; Wang, C.; Qin, Q.; Zheng, X.; Sun, Y. Temporal scale optimization for assimilation of spectral information and crop growth model. Trans. Chin. Soc. Agric. Eng. 2015, 142–148. [Google Scholar]
- Sakamoto, T.; Yokozawa, M.; Toritani, H.; Shibayama, M.; Ishitsuka, N.; Ohno, H. A crop phenology detection method using time-series MODIS data. Remote Sens. Environ. 2005, 96, 366–374. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Yao, X.; Deng, X.; Tian, Y.; Cao, W.; Zhu, Y. Detection of rice phenology through time series analysis of ground-based spectral index data. Field Crops Res. 2016, 198, 131–139. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, X.; Kou, W.; Qin, Y.; Zhang, G.; Li, L.; Jin, C.; Zhou, Y.; Wang, J.; Biradar, C. Tracking the dynamics of paddy rice planting area in 1986–2010 through time series Landsat images and phenology-based algorithms. Remote Sens. Environ. 2015, 160, 99–113. [Google Scholar] [CrossRef]
- Boogaard, H.L.; van Diepen, C.A.; Rutter, R.P.; Cabrera, J.M.C.A.; Laar, H.H. User’s Guide for the WOFOST 7.1 Crop Growth Simulation Model and WOFOST Control Center 1.5; Winand Staring Centre for Integrated Land, Soil and Water Research: Wageningen, The Netherlands, 1998. [Google Scholar]
- Onojeghuo, A.O.; Blackburn, G.A.; Wang, Q.; Atkinson, P.M.; Kindred, D.; Miao, Y. Rice crop phenology mapping at high spatial and temporal resolution using downscaled MODIS time-series. GISci. Remote Sens. 2018, 55, 659–677. [Google Scholar] [CrossRef]
- Peng, D.; Huete, A.R.; Huang, J.; Wang, F.; Sun, H. Detection and estimation of mixed paddy rice cropping patterns with MODIS data. Int. J. Appl. Earth Observ. Geoinform. 2011, 13, 13–23. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, Q.; Tan, J.; Zou, J. Regional scale mapping of fractional rice cropping change using a phenology-based temporal mixture analysis. Int. J. Remote Sens. 2018, 1–14. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, C.; Zeng, G.; Liang, J.; Guo, S.; Huang, L.; Wu, H.; Hua, S. Quantitative assessment of the contribution of climate variability and human activity to streamflow alteration in Dongting Lake, China. Hydrol. Process. 2016, 30, 1929–1939. [Google Scholar] [CrossRef]
- Qi, X.; Fu, Y.; Wang, R.Y.; Ng, C.N.; Dang, H.; He, Y. Improving the sustainability of agricultural land use: An integrated framework for the conflict between food security and environmental deterioration. Appl. Geogr. 2018, 90, 214–223. [Google Scholar] [CrossRef]
- Lu, R. Analytical Methods of Soil and Agricultural Chemistry; China Agricultural Science and Technology Press: Beijing, China, 1999; pp. 107–240. [Google Scholar]
- Didan, K.; Munoz, A.B.; Solano, R.; Huete, A. MODIS Vegetation Index User’s Guide (MOD13 Series); Vegetation Index and Phenology Lab., University of Arizona: Tucson, AZ, USA, 2015. [Google Scholar]
- Diepen, C.A.; Wolf, J.; Keulen, H.; Rappoldt, C. WOFOST: A simulation model of crop production. Soil Use Manag. 2010, 5, 16–24. [Google Scholar] [CrossRef]
- Wardlow, B.D.; Egbert, S.L. A comparison of MODIS 250-m EVI and NDVI data for crop mapping: A case study for southwest Kansas. Int. J. Remote Sens. 2010, 31, 805–830. [Google Scholar] [CrossRef]
- Fan, L.; Luo, Z.Z. Rice yield estimation based on MOSIS-NDVI: Case study of Chongqing Three Gorges warehouse district. Southwest China J. Agric. Sci. 2009, 1416–1419. [Google Scholar]
- Chen, P.Y.; Fedosejevs, G.; Tiscareño-López, M.; Arnold, J.G. Assessment of MODIS-EVI, MODIS-NDVI and VEGETATION-NDVI Composite Data Using Agricultural Measurements: An Example at Corn Fields in Western Mexico. Environ. Monit. Assess. 2006, 119, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, C.; Alfredo, H. From AVHRR-NDVI to MODIS-EVI: Advances in vegetation index research. Acta Ecol. Sin. 2003, 23, 979–987. [Google Scholar]
- Jönsson, P.; Eklundh, L. TIMESAT—A program for analyzing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef]
- Jonsson, P.; Eklundh, L. Seasonality extraction by function fitting to time-series of satellite sensor data. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- Li, S.H.; Xiao, J.T.; Ni, P.; Zhang, J.; Wang, H.S.; Wang, J.X. Monitoring paddy rice phenology using time series MODIS data over Jiangxi Province, China. Int. J. Agric. Biol. Eng. 2014, 7, 28–36. [Google Scholar]
- You, X.; Meng, J.; Zhang, M.; Dong, T. Remote Sensing Based Detection of Crop Phenology for Agricultural Zones in China Using a New Threshold Method. Remote Sens. 2013, 5, 3190–3211. [Google Scholar] [CrossRef]
- Yu, F.; Price, K.P.; Ellis, J.; Shi, P. Response of seasonal vegetation development to climatic variations in eastern central Asia. Remote Sens. Environ. 2003, 87, 42–54. [Google Scholar] [CrossRef]


| Different Strategies | Accuracy (R) a | Computation Time (h) | ||
|---|---|---|---|---|
| DOY203 | DOY219 | DOY236 | ||
| Assimilation with Phenology | 0.793 | 0.822 | 0.813 | 3.8 |
| Assimilation with EVI (8 images) | 0.703 | 0.714 | 0.722 | 8.4 |
| Assimilation with EVI (10 images) | 0.741 | 0.760 | 0.758 | 9.2 |
| Assimilation with EVI (12 images) | 0.795 | 0.824 | 0.810 | 10.7 |
| Assimilation with EVI (14 images) | 0.799 | 0.818 | 0.814 | 12.8 |
| Assimilation with phenology and EVI (12 images) | 0.80 | 0.825 | 0.816 | 13.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, G.; Liu, X.; Liu, M. Assimilating Remote Sensing Phenological Information into the WOFOST Model for Rice Growth Simulation. Remote Sens. 2019, 11, 268. https://doi.org/10.3390/rs11030268
Zhou G, Liu X, Liu M. Assimilating Remote Sensing Phenological Information into the WOFOST Model for Rice Growth Simulation. Remote Sensing. 2019; 11(3):268. https://doi.org/10.3390/rs11030268
Chicago/Turabian StyleZhou, Gaoxiang, Xiangnan Liu, and Ming Liu. 2019. "Assimilating Remote Sensing Phenological Information into the WOFOST Model for Rice Growth Simulation" Remote Sensing 11, no. 3: 268. https://doi.org/10.3390/rs11030268
APA StyleZhou, G., Liu, X., & Liu, M. (2019). Assimilating Remote Sensing Phenological Information into the WOFOST Model for Rice Growth Simulation. Remote Sensing, 11(3), 268. https://doi.org/10.3390/rs11030268