A Convolutional Neural Network Classifier Identifies Tree Species in Mixed-Conifer Forest from Hyperspectral Imagery
Abstract
:1. Introduction
1.1. Background and Problem
1.2. Convolutional Neural Networks
1.3. Research Objectives
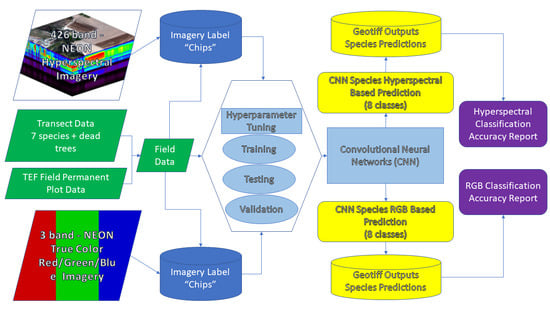
- Evaluate the application of CNNs to identify tree species in hyperspectral imagery compared to a Red-Green-Blue (RGB) subset of the hyperspectral imagery. We expected improved ability to accurately classify trees species using hyperspectral versus RGB imagery.
- Assess the accuracy of the tree species classification using a test dataset which is distinct from the training and validation data.
- Demonstrate potential uses of high-resolution tree species maps, i.e., analyze the distribution of trees across an elevation gradient.
- Provide tools so that other geospatial scientists can apply such techniques more broadly and evaluate the computational challenges to upscaling to larger areas.
2. Materials and Methods
2.1. Study Site and Airborne Imaging Data
2.2. Field Data and Study Species
2.3. Label Data Preparation for CNN Classification
2.4. CNN Model Architecture
2.5. Optimization/Hyperparameter Tuning/Prediction/Assessment
3. Results
3.1. CNN Classification and Parameter Settings
3.2. Application of High-Resolution Tree Species Mapping
4. Discussion
4.1. CNN Performance Using Hyperspectral Versus RGB Data
4.2. CNNs Versus Other Machine Learning Methods for Tree Species Identification
4.3. The Potential Uses of High-Resolution Tree Species Maps
4.4. Challenges in Computation and Upscaling to Broader Geographic Areas
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Abbreviation | Name | Description |
|---|---|---|
| AOP | NEON’s Airborne Observation Platform | A remote sensing system composed of an orthophoto camera, LiDAR sensor, and hyperspectral imager. |
| CHM | Canopy Height Model | The canopy height model was used to determine individual tree canopies |
| CNN | Convolutional Neural Network | The classification technique used to predict tree species from remote sensing imagery. This is also called ‘deep learning’ in the text. |
| DEM | Digital Elevation Model | Last return LiDAR derived surface representing the ground surface in our analysis |
| DSM | Digital Surface Model | The First-return LiDAR derived surface representing the tree canopy surface in our analysis |
| LiDAR | Light Detection and Ranging | The three-dimensional (3D) ranging technology used to measure the CHM, DSM, and DEM. |
| NEON | National Ecological Observatory Network | NEON was responsible for collecting the airborne remote sensing data in 2013. |
| SMA | Spectral Mixing Analysis | Classification method used to discriminate from multiple different spectra in pixels, often used with high spectral resolution imagery |
Appendix B. Instructions on How to Use the ‘Tree-Classification’ Toolkit
- Hyperspectral Imagery: data/NEON_D17_TEAK_DP1_20170627_181333_reflectance.tif
- Red-Green-Blue Imagery: data/NEON_D17_TEAK_DP1_20170627_181333_RGB_reflectance.tifCanopy height model: data/D17_CHM_all.tif
- Labels shapefile: data/Labels_Trimmed_Selective.shp
References
- Condit, R.; Ashton, P.S.; Baker, P.; Bunyavejchewin, S.; Gunatilleke, S.; Gunatilleke, N.; Hubbell, S.P.; Foster, R.B.; Itoh, A.; LaFrankie, J.V.; et al. Spatial Patterns in the Distribution of Tropical Tree Species. Science 2000, 288, 1414–1418. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.E.; Newman, S.D.; Aber, J.D.; Congalton, R.G. Determining forest species composition using high spectral resolution remote sensing data. Remote Sens. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Nagendra, H. Using remote sensing to assess biodiversity. Int. J. Remote Sens. 2001, 22, 2377–2400. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Mapping tree species in tropical seasonal semi-deciduous forests with hyperspectral and multispectral data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- Féret, J.-B.; Asner, G.P. Tree species discrimination in tropical forests using airborne imaging spectroscopy. IEEE Trans. Geosci. Remote Sens. 2012, 51, 73–84. [Google Scholar] [CrossRef]
- Colgan, M.; Baldeck, C.; Féret, J.-B.; Asner, G. Mapping savanna tree species at ecosystem scales using support vector machine classification and BRDF correction on airborne hyperspectral and LiDAR data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef]
- Laybros, A.; Schläpfer, D.; Feret, J.-B.; Descroix, L.; Bedeau, C.; Lefevre, M.-J.; Vincent, G. Across Date Species Detection Using Airborne Imaging Spectroscopy. Remote Sens. 2019, 11, 789. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual tree crown segmentation and classification of 13 tree species using Airborne hyperspectral data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Hyyppä, J.; Hyyppä, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int. J. Remote Sens. 2008, 29, 1339–1366. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Spies, T.A. An evaluation of alternate remote sensing products for forest inventory, monitoring, and mapping of Douglas-fir forests in western Oregon. Can. J. For. Res. 2001, 31, 78–87. [Google Scholar] [CrossRef]
- Tomppo, E.; Olsson, H.; Ståhl, G.; Nilsson, M.; Hagner, O.; Katila, M. Combining national forest inventory field plots and remote sensing data for forest databases. Remote Sens. Environ. 2008, 112, 1982–1999. [Google Scholar] [CrossRef]
- Wulder, M. Optical remote-sensing techniques for the assessment of forest inventory and biophysical parameters. Prog. Phys. Geogr. 1998, 22, 449–476. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J.; Im, J. Synergistic use of QuickBird multispectral imagery and LIDAR data for object-based forest species classification. Remote Sens. Environ. 2010, 114, 1141–1154. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Jones, M.O.; Martin, R.E.; Boardman, J.; Field, C.B. Carnegie Airborne Observatory: In-flight fusion of hyperspectral imaging and waveform light detection and ranging for three-dimensional studies of ecosystems. J. Appl. Remote Sens. 2007, 1, 13521–13536. [Google Scholar] [CrossRef]
- Marrs, J.; Ni-Meister, W. Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data. Remote Sens. 2019, 11, 819. [Google Scholar] [CrossRef]
- Pearlman, J.S.; Barry, P.S.; Segal, C.C.; Shepanski, J.; Beiso, D.; Carman, S.L. Hyperion, a space-based imaging spectrometer. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1160–1173. [Google Scholar] [CrossRef]
- Cudahy, T.J.; Hewson, R.; Huntington, J.F.; Quigley, M.A.; Barry, P.S. The performance of the satellite-borne Hyperion Hyperspectral VNIR-SWIR imaging system for mineral mapping at Mount Fitton, South Australia. In Proceedings of the IEEE 2001 International Geoscience and Remote Sensing Symposium (IGARSS’01), Sydney, Australia, 9–13 July 2001; Volume 1, pp. 314–316. [Google Scholar]
- Wettle, M.; Brando, V.E.; Dekker, A.G. A methodology for retrieval of environmental noise equivalent spectra applied to four Hyperion scenes of the same tropical coral reef. Remote Sens. Environ. 2004, 93, 188–197. [Google Scholar] [CrossRef]
- Pengra, B.W.; Johnston, C.A.; Loveland, T.R. Mapping an invasive plant, Phragmites australis, in coastal wetlands using the EO-1 Hyperion hyperspectral sensor. Remote Sens. Environ. 2007, 108, 74–81. [Google Scholar] [CrossRef]
- Kruse, F.A.; Boardman, J.W.; Huntington, J.F. Comparison of airborne hyperspectral data and EO-1 Hyperion for mineral mapping. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1388–1400. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sens. Environ. 2008, 112, 3958–3970. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2008, 7, 269–276. [Google Scholar] [CrossRef]
- Asner, G.P.; Hughes, R.F.; Vitousek, P.M.; Knapp, D.E.; Kennedy-Bowdoin, T.; Boardman, J.; Martin, R.E.; Eastwood, M.; Green, R.O. Invasive plants transform the three-dimensional structure of rain forests. Proc. Natl. Acad. Sci. USA 2008, 105, 4519–4523. [Google Scholar] [CrossRef] [Green Version]
- Doughty, C.; Asner, G.; Martin, R. Predicting tropical plant physiology from leaf and canopy spectroscopy. Oecologia 2011, 165, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Kampe, T.U.; Johnson, B.R.; Kuester, M.A.; Keller, M. NEON: The first continental-scale ecological observatory with airborne remote sensing of vegetation canopy biochemistry and structure. Int. Soc. Opt. Eng. 2010, 4, 43510–43524. [Google Scholar] [CrossRef]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N. Individual tree detection and classification with UAV-based photogrammetric point clouds and hyperspectral imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef]
- Tuominen, S.; Näsi, R.; Honkavaara, E.; Balazs, A.; Hakala, T.; Viljanen, N.; Pölönen, I.; Saari, H.; Ojanen, H. Assessment of classifiers and remote sensing features of hyperspectral imagery and stereo-photogrammetric point clouds for recognition of tree species in a forest area of high species diversity. Remote Sens. 2018, 10, 714. [Google Scholar] [CrossRef]
- Sothe, C.; Dalponte, M.; de Almeida, C.M.; Schimalski, M.B.; Lima, C.L.; Liesenberg, V.; Miyoshi, G.T.; Tommaselli, A.M.G. Tree Species Classification in a Highly Diverse Subtropical Forest Integrating UAV-Based Photogrammetric Point Cloud and Hyperspectral Data. Remote Sens. 2019, 11, 1338. [Google Scholar] [CrossRef]
- Shen, X.; Cao, L. Tree-species classification in subtropical forests using airborne hyperspectral and LiDAR data. Remote Sens. 2017, 9, 1180. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Dalponte, M.; Ene, L.T.; Marconcini, M.; Gobakken, T.; Næsset, E. Semi-supervised SVM for individual tree crown species classification. ISPRS J. Photogramm. Remote Sens. 2015, 110, 77–87. [Google Scholar] [CrossRef]
- Graves, S.; Asner, G.; Martin, R.; Anderson, C.; Colgan, M.; Kalantari, L.; Bohlman, S. Tree species abundance predictions in a tropical agricultural landscape with a supervised classification model and imbalanced data. Remote Sens. 2016, 8, 161. [Google Scholar] [CrossRef]
- Zhang, C.; Qiu, F. Mapping Individual Tree Species in an Urban Forest Using Airborne Lidar Data and Hyperspectral Imagery. Photogramm. Eng. Remote Sens. 2013, 78, 1079–1087. [Google Scholar] [CrossRef]
- Pu, R. Broadleaf species recognition with in situ hyperspectral data. Int. J. Remote Sens. 2009, 30, 2759–2779. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Stateline, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef] [PubMed]
- Ciresan, D.C.; Meier, U.; Gambardella, L.M.; Schmidhuber, J. Convolutional Neural Network Committees for Handwritten Character Classification. In Proceedings of the 2011 International Conference on Document Analysis and Recognition, Washington, DC, USA, 18–21 September 2011; pp. 1135–1139. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Cireşan, D.; Meier, U.; Masci, J.; Schmidhuber, J. Multi-column deep neural network for traffic sign classification. Neural Netw. 2012, 32, 333–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y. Convolutional neural networks for sentence classification. arXiv 2014, arXiv:1408.5882. [Google Scholar]
- Lawrence, S.; Giles, C.L.; Ah Chung, T.; Back, A.D. Face recognition: A convolutional neural-network approach. IEEE Trans. Neural Netw. 1997, 8, 98–113. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Chan, C.S.; Wilkin, P.; Remagnino, P. Deep-plant: Plant identification with convolutional neural networks. Proceedings of 2015 IEEE International Conference on the Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 452–456. [Google Scholar]
- Norouzzadeh, M.S.; Nguyen, A.; Kosmala, M.; Swanson, A.; Palmer, M.S.; Packer, C.; Clune, J. Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proc. Natl. Acad. Sci. USA 2018, 115, E5716–E5725. [Google Scholar] [CrossRef] [Green Version]
- Van Horn, G.; Mac Aodha, O.; Song, Y.; Shepard, A.; Adam, H.; Perona, P.; Belongie, S. The inaturalist challenge 2017 dataset. arXiv 2017, arXiv:1707.06642. [Google Scholar]
- Bergstra, J.; Bastien, F.; Breuleux, O.; Lamblin, P.; Pascanu, R.; Delalleau, O.; Desjardins, G.; Warde-Farley, D.; Goodfellow, I.; Bergeron, A. Theano: Deep learning on gpus with python. In Proceedings of the NIPS 2011, BigLearning Workshop, Granada, Spain, 16–17 December 2011; Microtome Publishing: Brookline, MA, USA, 2011; Volume 3, pp. 1–48. [Google Scholar]
- Woodcock, C.E.; Strahler, A.H. The factor of scale in remote sensing. Remote Sens. Environ. 1987, 21, 311–332. [Google Scholar] [CrossRef]
- Castelluccio, M.; Poggi, G.; Sansone, C.; Verdoliva, L. Land use classification in remote sensing images by convolutional neural networks. arXiv 2015, arXiv:1508.00092. [Google Scholar]
- Zhang, Q.; Yuan, Q.; Zeng, C.; Li, X.; Wei, Y. Missing Data Reconstruction in Remote Sensing Image with a Unified Spatial-Temporal-Spectral Deep Convolutional Neural Network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4274–4288. [Google Scholar] [CrossRef]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef]
- Hu, F.; Xia, G.-S.; Hu, J.; Zhang, L. Transferring Deep Convolutional Neural Networks for the Scene Classification of High-Resolution Remote Sensing Imagery. Remote Sens. 2015, 7, 14680–14707. [Google Scholar] [CrossRef] [Green Version]
- Penatti, O.A.B.; Nogueira, K.; Dos Santos, J.A. Do deep features generalize from everyday objects to remote sensing and aerial scenes domains? In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Boston, MA, USA, 7–12 June 2015; pp. 44–51. [Google Scholar]
- Cheng, G.; Yang, C.; Yao, X.; Guo, L.; Han, J. When deep learning meets metric learning: Remote sensing image scene classification via learning discriminative CNNs. IEEE Trans. Geosci. Remote Sens. 2018, 56, 2811–2821. [Google Scholar] [CrossRef]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep convolutional neural networks for hyperspectral image classification. J. Sens. 2015, 2015, 258619. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Kampe, T.; Leisso, N.; Musinsky, J.; Petroy, S.; Karpowiez, B.; Krause, K.; Crocker, R.I.; DeVoe, M.; Penniman, E.; Guadagno, T. The NEON 2013 airborne campaign at domain 17 terrestrial and aquatic sites in California; NEON Technical Memorandum Service TM 005: Boulder, CO, USA, 2013. [Google Scholar]
- North, M.; Oakley, B.; Chen, J.; Erickson, H.; Gray, A.; Izzo, A.; Johnson, D.; Ma, S.; Marra, J.; Meyer, M. Vegetation and Ecological Characteristics of Mixed-Conifer and Red Fir Forests at the Teakettle Experimental Forest; Technical Report PSW-GTR-186, Pacific Southwest Research Station, Forest Service US Department. Agriculture: Albany, CA, USA, 2002; Volume 186, p. 52. [Google Scholar]
- Lucas, R.; Armston, J.; Fairfax, R.; Fensham, R.; Accad, A.; Carreiras, J.; Kelley, J.; Bunting, P.; Clewley, D.; Bray, S. An evaluation of the ALOS PALSAR L-band backscatter—Above ground biomass relationship Queensland, Australia: Impacts of surface moisture condition and vegetation structure. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2010, 3, 576–593. [Google Scholar] [CrossRef]
- Cartus, O.; Santoro, M.; Kellndorfer, J. Mapping forest aboveground biomass in the Northeastern United States with ALOS PALSAR dual-polarization L-band. Remote Sens. Environ. 2012, 124, 466–478. [Google Scholar] [CrossRef]
- Evans, D.L.; Carraway, R.W.; Simmons, G.T. Use of Global Positioning System (GPS) for Forest Plot Location. South. J. Appl. For. 1992, 16, 67–70. [Google Scholar] [CrossRef]
- Popescu, S.C. Estimating biomass of individual pine trees using airborne lidar. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Barry, S.; Elith, J. Error and uncertainty in habitat models. J. Appl. Ecol. 2006, 43, 413–423. [Google Scholar] [CrossRef]
- Meyer, M.D.; North, M.P.; Gray, A.N.; Zald, H.S.J. Influence of soil thickness on stand characteristics in a Sierra Nevada mixed-conifer forest. Plant Soil 2007, 294, 113–123. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Stehman, S.V. Selecting and interpreting measures of thematic classification accuracy. Remote Sens. Environ. 1997, 62, 77–89. [Google Scholar] [CrossRef]
- Peet, R.K. The measurement of species diversity. Annu. Rev. Ecol. Syst. 1974, 5, 285–307. [Google Scholar] [CrossRef]
- Urban, D.L.; Miller, C.; Halpin, P.N.; Stephenson, N.L. Forest gradient response in Sierran landscapes: The physical template. Landsc. Ecol. 2000, 15, 603–620. [Google Scholar] [CrossRef]
- North, M.; Collins, B.M.; Safford, H.; Stephenson, N.L. Montane Forests; Mooney, H., Zavaleta, E., Eds.; University of California Press: Berkeley, CA, USA, 2016; Chapter 27; pp. 553–577. [Google Scholar]
- Garrido-Novell, C.; Pérez-Marin, D.; Amigo, J.M.; Fernández-Novales, J.; Guerrero, J.E.; Garrido-Varo, A. Grading and color evolution of apples using RGB and hyperspectral imaging vision cameras. J. Food Eng. 2012, 113, 281–288. [Google Scholar] [CrossRef]
- Okamoto, H.; Murata, T.; Kataoka, T.; HATA, S. Plant classification for weed detection using hyperspectral imaging with wavelet analysis. Weed Biol. Manag. 2007, 7, 31–37. [Google Scholar] [CrossRef]
- Bauriegel, E.; Giebel, A.; Geyer, M.; Schmidt, U.; Herppich, W.B. Early detection of Fusarium infection in wheat using hyper-spectral imaging. Comput. Electron. Agric. 2011, 75, 304–312. [Google Scholar] [CrossRef]
- Makantasis, K.; Karantzalos, K.; Doulamis, A.; Doulamis, N. Deep supervised learning for hyperspectral data classification through convolutional neural networks. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 4959–4962. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin, Germany; pp. 234–241. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
- Gong, P.; Pu, R.; Yu, B. Conifer species recognition: An exploratory analysis of in situ hyperspectral data. Remote Sens. Environ. 1997, 62, 189–200. [Google Scholar] [CrossRef]
- Asner, G.P.; Brodrick, P.G.; Anderson, C.B.; Vaughn, N.; Knapp, D.E.; Martin, R.E. Progressive forest canopy water loss during the 2012–2015 California drought. Proc. Natl. Acad. Sci. USA 2016, 113, E249. [Google Scholar] [CrossRef] [PubMed]
- Hawes, J.E.; Peres, C.A.; Riley, L.B.; Hess, L.L. Landscape-scale variation in structure and biomass of Amazonian seasonally flooded and unflooded forests. For. Ecol. Manag. 2012, 281, 163–176. [Google Scholar] [CrossRef]
- He, H.S.; Mladenoff, D.J. Spa tially explicit and stochastic simulation of forest-landscape fire disturbance and succession. Ecology 1999, 80, 81–99. [Google Scholar] [CrossRef]
- Conroy, M.J.; Noon, B.R. Mapping of Species Richness for Conservation of Biological Diversity: Conceptual and Methodological Issues. Ecol. Appl. 1996, 6, 763–773. [Google Scholar] [CrossRef]
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.H.T. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef] [Green Version]
- Brell, M.; Segl, K.; Guanter, L.; Bookhagen, B. Hyperspectral and lidar intensity data fusion: A framework for the rigorous correction of illumination, anisotropic effects, and cross calibration. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2799–2810. [Google Scholar] [CrossRef]
- Schimel, D.S.; Asner, G.P.; Moorcroft, P. Observing changing ecological diversity in the Anthropocene. Front. Ecol. Environ. 2013, 11, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Das, A.J.; Stephenson, N.L.; Flint, A.; Das, T.; Van Mantgem, P.J. Climatic correlates of tree mortality in water-and energy-limited forests. PLoS ONE 2013, 8, e69917. [Google Scholar] [CrossRef]
- Allen, C.D.; Breshears, D.D.; McDowell, N.G. On underestimation of global vulnerability to tree mortality and forest die-off from hotter drought in the Anthropocene. Ecosphere 2015, 6, 1–55. [Google Scholar] [CrossRef]
- Underwood, E.C.; Mulitsch, M.J.; Greenberg, J.A.; Whiting, M.L.; Ustin, S.L.; Kefauver, S.C. Mapping Invasive Aquatic Vegetation in the Sacramento-San Joaquin Delta using Hyperspectral Imagery. Environ. Monit. Assess. 2006, 121, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Korpela, I.; Heikkinen, V.; Honkavaara, E.; Rohrbach, F.; Tokola, T. Variation and directional anisotropy of reflectance at the crown scale—Implications for tree species classification in digital aerial images. Remote Sens. Environ. 2011, 115, 2062–2074. [Google Scholar] [CrossRef]
- Chang, C.-I.; Du, Q. Estimation of number of spectrally distinct signal sources in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 608–619. [Google Scholar] [CrossRef]
- Chen, G.; Qian, S.-E. Denoising of hyperspectral imagery using principal component analysis and wavelet shrinkage. IEEE Trans. Geosci. Remote Sens. 2011, 49, 973–980. [Google Scholar] [CrossRef]
- Culvenor, D.S. Extracting individual tree information. In Remote Sensing of Forest Environments; Springer: Berlin, Germany, 2003; pp. 255–277. [Google Scholar]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar detection of individual tree size in tropical forests. Remote Sens. Environ. 2016, 183, 318–333. [Google Scholar] [CrossRef]
- Edwards, T.C., Jr.; Cutler, D.R.; Zimmermann, N.E.; Geiser, L.; Moisen, G.G. Effects of sample survey design on the accuracy of classification tree models in species distribution models. Ecol. Model. 2006, 199, 132–141. [Google Scholar] [CrossRef]





| Code | Scientific Name (Common Name) | Abbreviation | Number |
|---|---|---|---|
| 0 | Abies concolor (White fir) | abco | 119 |
| 1 | Abies magnifica (Red fir) | abma | 47 |
| 2 | Calocedrus decurrens (Incense cedar) | cade | 66 |
| 3 | Pinus jeffreyi (Jeffrey pine) | pije | 164 |
| 4 | Pinus lambertiana (Sugar pine) | pila | 68 |
| 5 | Quercus kelloggii (Black oak) | quke | 18 |
| 6 | Pinus contorta (Lodgepole pine) | pico | 62 |
| 7 | Dead (any species) | dead | 169 |
| Species | Species Code | Hyperspectral | RGB | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-Score | ||
| White fir | 0 | 0.76 | 0.81 | 0.78 | 0.46 | 0.53 | 0.49 |
| Red fir | 1 | 0.76 | 0.72 | 0.74 | 0.41 | 0.30 | 0.35 |
| Incense cedar | 2 | 0.90 | 0.85 | 0.88 | 0.50 | 0.44 | 0.47 |
| Jeffrey pine | 3 | 0.93 | 0.96 | 0.95 | 0.65 | 0.73 | 0.69 |
| Sugar pine | 4 | 0.90 | 0.96 | 0.93 | 0.67 | 0.68 | 0.67 |
| Black oak | 5 | 0.73 | 0.61 | 0.67 | 0.69 | 0.61 | 0.65 |
| Lodgepole pine | 6 | 0.84 | 0.87 | 0.86 | 0.54 | 0.47 | 0.50 |
| Dead | 7 | 0.90 | 0.85 | 0.88 | 0.88 | 0.86 | 0.87 |
| Ave/Total | 0.87 | 0.87 | 0.87 | 0.64 | 0.64 | 0.64 | |
| Species | Abbrev | Spp Code | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Recall | F-Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0. White fir | abma | 0 | 96 | 7 | 2 | 2 | 2 | 0 | 1 | 9 | 0.81 | 0.78 |
| 1. Red fir | abco | 1 | 11 | 34 | 1 | 0 | 0 | 0 | 0 | 1 | 0.72 | 0.74 |
| 2. Incense Cedar | cade | 2 | 3 | 0 | 56 | 3 | 0 | 0 | 0 | 4 | 0.85 | 0.88 |
| 3. Jeffrey pine | pije | 3 | 1 | 0 | 2 | 158 | 1 | 0 | 1 | 1 | 0.96 | 0.95 |
| 4. Sugar pine | pila | 4 | 1 | 0 | 1 | 1 | 65 | 0 | 0 | 0 | 0.96 | 0.93 |
| 5. Black oak | quke | 5 | 0 | 0 | 0 | 0 | 0 | 11 | 6 | 1 | 0.61 | 0.67 |
| 6. Lodgepole pine | pico | 6 | 0 | 0 | 0 | 2 | 2 | 4 | 54 | 0 | 0.87 | 0.86 |
| 7. Dead | dead | 7 | 14 | 4 | 0 | 3 | 2 | 0 | 2 | 144 | 0.85 | 0.88 |
| Precision | 0.76 | 0.76 | 0.90 | 0.93 | 0.90 | 0.73 | 0.84 | 0.90 |
| Species | Abbrev | Spp Code | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Recall |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0. White fir | abma | 0 | 63 | 11 | 9 | 17 | 3 | 0 | 5 | 11 | 0.53 |
| 1. Red fir | abco | 1 | 16 | 14 | 3 | 9 | 1 | 0 | 2 | 2 | 0.30 |
| 2. Incense Cedar | cade | 2 | 15 | 2 | 29 | 9 | 3 | 1 | 6 | 1 | 0.44 |
| 3. Jeffrey pine | pije | 3 | 16 | 3 | 5 | 119 | 12 | 1 | 5 | 3 | 0.73 |
| 4. Sugar pine | pila | 4 | 3 | 1 | 4 | 12 | 46 | 0 | 2 | 0 | 0.68 |
| 5. Black oak | quke | 5 | 0 | 0 | 0 | 1 | 1 | 11 | 4 | 1 | 0.61 |
| 6. Lodgepole pine | pico | 6 | 14 | 1 | 4 | 9 | 2 | 2 | 29 | 1 | 0.47 |
| 7. Dead | dead | 7 | 9 | 2 | 4 | 6 | 1 | 1 | 1 | 145 | 0.86 |
| Precision | 0.46 | 0.41 | 0.50 | 0.65 | 0.67 | 0.69 | 0.54 | 0.88 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A Convolutional Neural Network Classifier Identifies Tree Species in Mixed-Conifer Forest from Hyperspectral Imagery. Remote Sens. 2019, 11, 2326. https://doi.org/10.3390/rs11192326
Fricker GA, Ventura JD, Wolf JA, North MP, Davis FW, Franklin J. A Convolutional Neural Network Classifier Identifies Tree Species in Mixed-Conifer Forest from Hyperspectral Imagery. Remote Sensing. 2019; 11(19):2326. https://doi.org/10.3390/rs11192326
Chicago/Turabian StyleFricker, Geoffrey A., Jonathan D. Ventura, Jeffrey A. Wolf, Malcolm P. North, Frank W. Davis, and Janet Franklin. 2019. "A Convolutional Neural Network Classifier Identifies Tree Species in Mixed-Conifer Forest from Hyperspectral Imagery" Remote Sensing 11, no. 19: 2326. https://doi.org/10.3390/rs11192326