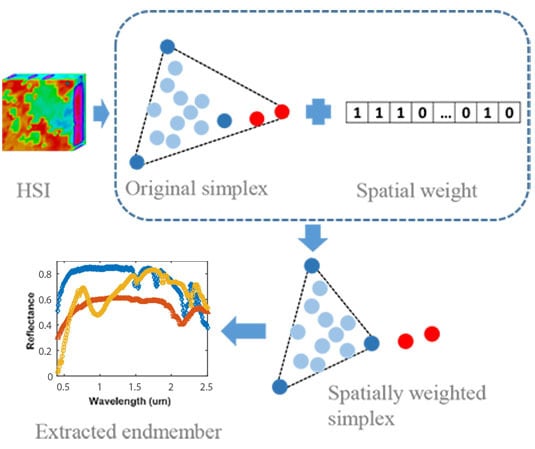
Figure 1.
Illustration of the SWSS. As we can see from this image, HSI is displayed in (a); According to the HSI, the SWSS defines the spatial weight scalar (c) for each pixel within the HSI using a value of 0 or 1. Then, the spatial weight is used to weight the pixel within the simplex, where the light blue, dark blue, and red spheres denote the mixed pixel, endmember, and anomalies, respectively (b); Based on the spatially weighed simplex, the vertices of the simplex, i.e., endmembers, are updated and the anomalies avoided (d); Finally, the extracted endmembers are displayed in (e).
Figure 1.
Illustration of the SWSS. As we can see from this image, HSI is displayed in (a); According to the HSI, the SWSS defines the spatial weight scalar (c) for each pixel within the HSI using a value of 0 or 1. Then, the spatial weight is used to weight the pixel within the simplex, where the light blue, dark blue, and red spheres denote the mixed pixel, endmember, and anomalies, respectively (b); Based on the spatially weighed simplex, the vertices of the simplex, i.e., endmembers, are updated and the anomalies avoided (d); Finally, the extracted endmembers are displayed in (e).
Figure 2.
Visual illustration of the endmember extraction process of the simplex. For the extreme projection-based simplex, an endmember is identified by its extreme projection into a subspace (a); For the maximum simplex volume-based simplex, endmembers are related to the pixels that can define a maximum simplex volume (b). In these two figures, light spheres and the dark spheres denote the mixed pixels and the determined endmembers, respectively.
Figure 2.
Visual illustration of the endmember extraction process of the simplex. For the extreme projection-based simplex, an endmember is identified by its extreme projection into a subspace (a); For the maximum simplex volume-based simplex, endmembers are related to the pixels that can define a maximum simplex volume (b). In these two figures, light spheres and the dark spheres denote the mixed pixels and the determined endmembers, respectively.
Figure 3.
Synthetic data set without anomalies. In this synthetic hyperspectral image, it contains 224 bands, and the image is displayed in the top-left position. Five endmembers, namely Alunite SUSTDA-20, Calcite HS48.3B, Kaolin/Smect KLF506 95%K, Montmorillonite STx-1, and Muscovite GDS111 Guatemal, are shown in the top-right position, and their corresponding abundance maps are displayed in the bottom position.
Figure 3.
Synthetic data set without anomalies. In this synthetic hyperspectral image, it contains 224 bands, and the image is displayed in the top-left position. Five endmembers, namely Alunite SUSTDA-20, Calcite HS48.3B, Kaolin/Smect KLF506 95%K, Montmorillonite STx-1, and Muscovite GDS111 Guatemal, are shown in the top-right position, and their corresponding abundance maps are displayed in the bottom position.
Figure 4.
Synthetic hyperspectral image without anomalies (left) and with anomalies (right). Both synthetic images are comprised of multiple blocks filled by mixed pixels with different spectral purities: i.e., 1 (pure pixels ), 0.8 (mixed with 2 materials), 0.6 (mixed with 3 materials), and 0.4 (mixed with 4 materials). The background of the image was made up of mixed pixels averaged on all endmembers (the purity of each material was 0.2). In addition, for the synthetic image with anomalies (right), it contains different anomaly areas. Five panels of different sizes are added to the image, and the sizes are 1 × 1, 2 × 2, 2 × 3, 3 × 3, and 3 × 5.
Figure 4.
Synthetic hyperspectral image without anomalies (left) and with anomalies (right). Both synthetic images are comprised of multiple blocks filled by mixed pixels with different spectral purities: i.e., 1 (pure pixels ), 0.8 (mixed with 2 materials), 0.6 (mixed with 3 materials), and 0.4 (mixed with 4 materials). The background of the image was made up of mixed pixels averaged on all endmembers (the purity of each material was 0.2). In addition, for the synthetic image with anomalies (right), it contains different anomaly areas. Five panels of different sizes are added to the image, and the sizes are 1 × 1, 2 × 2, 2 × 3, 3 × 3, and 3 × 5.
Figure 5.
(a–t) Visual comparison between the spectra library (blue solid line) and the extracted endmembers (the red and yellow solid lines denote the original and improved versions, respectively) when the experiments were conducted on the synthetic image with 40 dB and the was fixed to 7 (without the interference of anomalies): for (a–d) Alunite SUSTDA-20; (e–h) Calcite HS48.3B; (i–l) Kaolin/Smect KLF506 95%K; (m–p) Montmorillonite STx-1; and (q–t) Muscovite GDS111 Guatemal; (a–q) OSP and SWSS-OSP; (b–r) N-FINDR and SWSS-N-FINDR; (c–s) VCA and SWSS-VCA; (d–t) AVMAX and SWSS-AVMAX.
Figure 5.
(a–t) Visual comparison between the spectra library (blue solid line) and the extracted endmembers (the red and yellow solid lines denote the original and improved versions, respectively) when the experiments were conducted on the synthetic image with 40 dB and the was fixed to 7 (without the interference of anomalies): for (a–d) Alunite SUSTDA-20; (e–h) Calcite HS48.3B; (i–l) Kaolin/Smect KLF506 95%K; (m–p) Montmorillonite STx-1; and (q–t) Muscovite GDS111 Guatemal; (a–q) OSP and SWSS-OSP; (b–r) N-FINDR and SWSS-N-FINDR; (c–s) VCA and SWSS-VCA; (d–t) AVMAX and SWSS-AVMAX.
Figure 6.
(a–t) Visual comparison between the spectra library (blue solid line) and the extracted endmembers (the red and yellow solid lines denote the original and improved versions, respectively) when the experiments were conducted on the synthetic image with 40 dB and the was fixed to 7 (with the interference of anomalies): for (a–d) Alunite SUSTDA-20; (e–h) Calcite HS48.3B; (i–l) Kaolin/Smect KLF506 95%K; (m–p) Montmorillonite STx-1; and (q–t) Muscovite GDS111 Guatemal; (a–q) OSP and SWSS-OSP; (b–r) N-FINDR and SWSS-N-FINDR; (c–s) VCA and SWSS-VCA; (d–t) AVMAX and SWSS-AVMAX.
Figure 6.
(a–t) Visual comparison between the spectra library (blue solid line) and the extracted endmembers (the red and yellow solid lines denote the original and improved versions, respectively) when the experiments were conducted on the synthetic image with 40 dB and the was fixed to 7 (with the interference of anomalies): for (a–d) Alunite SUSTDA-20; (e–h) Calcite HS48.3B; (i–l) Kaolin/Smect KLF506 95%K; (m–p) Montmorillonite STx-1; and (q–t) Muscovite GDS111 Guatemal; (a–q) OSP and SWSS-OSP; (b–r) N-FINDR and SWSS-N-FINDR; (c–s) VCA and SWSS-VCA; (d–t) AVMAX and SWSS-AVMAX.
Figure 7.
(a) Execution times of original versions, improved versions (with displayed in parentheses), and SPP coupled simplex-based algorithms for experiments conducted on synthetic dataset without anomalies; (b) execution times of original versions, improved versions (with displayed in parentheses), and SPP coupled simplex-based algorithms for experiments conducted on synthetic dataset with anomalies.
Figure 7.
(a) Execution times of original versions, improved versions (with displayed in parentheses), and SPP coupled simplex-based algorithms for experiments conducted on synthetic dataset without anomalies; (b) execution times of original versions, improved versions (with displayed in parentheses), and SPP coupled simplex-based algorithms for experiments conducted on synthetic dataset with anomalies.
Figure 8.
Joint consideration of different window size and noise scenarios. X axis denotes window size varying from 0 to 9, where equals 0 denotes original version. Y axis denotes noise scenarios varying from 10 to 60 dB. Z axis denotes SAD results yielded by algorithms under different combinations of window size and noise scenarios. (a–d) denote SAD results generated by SWSS-OSP, SWSS-N-FINDR, SWSS-VCA, and SWSS-AVMAX, respectively, on synthetic dataset without anomalies added; (e–h) denote SAD results generated by SWSS-OSP, SWSS-N-FINDR, SWSS-VCA, and SWSS-AVMAX, respectively, on synthetic dataset with anomalies added.
Figure 8.
Joint consideration of different window size and noise scenarios. X axis denotes window size varying from 0 to 9, where equals 0 denotes original version. Y axis denotes noise scenarios varying from 10 to 60 dB. Z axis denotes SAD results yielded by algorithms under different combinations of window size and noise scenarios. (a–d) denote SAD results generated by SWSS-OSP, SWSS-N-FINDR, SWSS-VCA, and SWSS-AVMAX, respectively, on synthetic dataset without anomalies added; (e–h) denote SAD results generated by SWSS-OSP, SWSS-N-FINDR, SWSS-VCA, and SWSS-AVMAX, respectively, on synthetic dataset with anomalies added.
Figure 10.
RMSE results of the original version and the improved version with different window sizes for experiments conducted on the dataset.
Figure 10.
RMSE results of the original version and the improved version with different window sizes for experiments conducted on the dataset.
Figure 11.
The location of the extracted endmembers. (a–d) Endmembers extracted from the original versions, i.e., OSP, N-FINDR, VCA, and AVMAX; and (e–h) endmembers extracted from the improved versions, i.e., SWSS-OSP, SWSS-N-FINDR, SWSS-VCA, and SWSS-AVMAX.
Figure 11.
The location of the extracted endmembers. (a–d) Endmembers extracted from the original versions, i.e., OSP, N-FINDR, VCA, and AVMAX; and (e–h) endmembers extracted from the improved versions, i.e., SWSS-OSP, SWSS-N-FINDR, SWSS-VCA, and SWSS-AVMAX.
Table 1.
The segmentation specified by the OTSU algorithm; it was used to segment the spatial information of each pixel of the synthetic image (without anomalies) under different SNRs and window sizes .
Table 1.
The segmentation specified by the OTSU algorithm; it was used to segment the spatial information of each pixel of the synthetic image (without anomalies) under different SNRs and window sizes .
| Window Size | ws = 3 | ws = 5 | ws = 7 | ws = 9 |
|---|
| 10 dB | 0.3647 | 0.4314 | 0.4549 | 0.4706 |
| 20 dB | 0.3686 | 0.4431 | 0.4353 | 0.4078 |
| 30 dB | 0.2863 | 0.3490 | 0.3804 | 0.3961 |
| 40 dB | 0.2510 | 0.3255 | 0.3647 | 0.3686 |
| 50 dB | 0.2196 | 0.3118 | 0.3314 | 0.3059 |
| 60 dB | 0.2588 | 0.3176 | 0.3255 | 0.3333 |
Table 2.
The segmentation threshold specified by the OTSU algorithm; it was used to segment the spatial information of each pixel of the synthetic image (with anomalies) under different SNRs and window sizes .
Table 2.
The segmentation threshold specified by the OTSU algorithm; it was used to segment the spatial information of each pixel of the synthetic image (with anomalies) under different SNRs and window sizes .
| Window Size | ws = 3 | ws = 5 | ws = 7 | ws = 9 |
|---|
| 10 dB | 0.4667 | 0.5294 | 0.5373 | 0.5294 |
| 20 dB | 0.4000 | 0.4549 | 0.4784 | 0.4588 |
| 30 dB | 0.2980 | 0.3804 | 0.3961 | 0.4157 |
| 40 dB | 0.2627 | 0.3333 | 0.3647 | 0.3725 |
| 50 dB | 0.2431 | 0.2784 | 0.3275 | 0.3353 |
| 60 dB | 0.2118 | 0.3137 | 0.3294 | 0.3431 |
Table 3.
The segmentation specified by the OTSU algorithm; it was used to segment the spatial information of each pixel on the dataset under different window sizes .
Table 3.
The segmentation specified by the OTSU algorithm; it was used to segment the spatial information of each pixel on the dataset under different window sizes .
| Window Size | ws = 3 | ws = 5 | ws = 7 | ws = 9 |
|---|
| 0.1882 | 0.1882 | 0.1804 | 0.1686 |
Table 4.
SAD results of the original and improved versions, with the experiment conducted on a synthetic image (without anomalies) under different SNRs.
Table 4.
SAD results of the original and improved versions, with the experiment conducted on a synthetic image (without anomalies) under different SNRs.
| Algorithms | OSP | SWSS-OSP | N-FINDR | SWSS-N-FINDR | VCA | SWSS-VCA | AVMAX | SWSS-AVMAX |
|---|
| 10 dB | 0.4327 | 0.4212 | 0.4170 | 0.4030 | 0.1166 | 0.0826 | 0.4199 | 0.3991 |
| 20 dB | 0.1475 | 0.1587 | 0.1417 | 0.1411 | 0.0316 | 0.0223 | 0.1399 | 0.1396 |
| 30 dB | 0.0457 | 0.0450 | 0.0453 | 0.0449 | 0.0092 | 0.0078 | 0.0448 | 0.0447 |
| 40 db | 0.0143 | 0.0143 | 0.0156 | 0.0150 | 0.0029 | 0.0026 | 0.0143 | 0.0141 |
| 50 dB | 0.0045 | 0.0044 | 0.0047 | 0.0045 | 0.009 | 0.0008 | 0.0045 | 0.0045 |
| 60 dB | 0.0014 | 0.0014 | 0.0018 | 0.0018 | 0.0003 | 0.0003 | 0.0014 | 0.0017 |
| Average | 0.1077 | 0.1081 | 0.1044 | 0.1011 | 0.0269 | 0.0201 | 0.1041 | 0.1006 |
Table 5.
SAD results of the original and improved versions for experiments conducted on a synthetic image (with anomalies) under different SNRs.
Table 5.
SAD results of the original and improved versions for experiments conducted on a synthetic image (with anomalies) under different SNRs.
| Algorithms | OSP | SWSS-OSP | N-FINDR | SWSS-N-FINDR | VCA | SWSS-VCA | AVMAX | SWSS-AVMAX |
|---|
| 10 dB | 0.4214 | 0.4212 | 0.4270 | 0.4030 | 0.1245 | 0.0826 | 0.4263 | 0.4044 |
| 20 dB | 0.1798 | 0.1543 | 0.1568 | 0.1386 | 0.0581 | 0.0213 | 0.1555 | 0.1385 |
| 30 dB | 0.0776 | 0.0456 | 0.0791 | 0.0444 | 0.0603 | 0.0074 | 0.0725 | 0.0442 |
| 40 db | 0.0625 | 0.0141 | 0.0537 | 0.0152 | 0.0558 | 0.0025 | 0.0557 | 0.0141 |
| 50 dB | 0.0553 | 0.0044 | 0.0465 | 0.0046 | 0.0555 | 0.0009 | 0.0546 | 0.0044 |
| 60 dB | 0.0549 | 0.0014 | 0.0527 | 0.0014 | 0.0548 | 0.0003 | 0.0509 | 0.0014 |
| Average | 0.1419 | 0.1068 | 0.1360 | 0.1011 | 0.0665 | 0.0192 | 0.1359 | 0.1012 |
Table 6.
SAD results and execution time (tabulated in parentheses) obtained from the improved versions under different window sizes and in two synthetic dataset scenarios.
Table 6.
SAD results and execution time (tabulated in parentheses) obtained from the improved versions under different window sizes and in two synthetic dataset scenarios.
| Window Size | ws = 3 | ws = 5 | ws = 7 | ws = 9 |
|---|
| SWSS-OSP | 0.1069 (6.7327) | 0.1081 (10.1887) | 0.1085 (14.1675) | 0.1087 (20.8113) |
| SWSS-N-FINDR | 0.1010 (4.3943) | 0.1003 (7.8081) | 0.1010 (12.3514) | 0.1021 (18.3483) |
| SWSS-VCA | 0.0204 (3.9102) | 0.0215 (7.3713) | 0.0178 (11.9351) | 0.0207 (17.9347) |
| SWSS-AVMAX | 0.1005 (3.6717) | 0.1006 (7.0336) | 0.1003 (11.6494) | 0.1011 (17.5922) |
| Average | 0.0822 (4.6772) | 0.0826 (8.1004) | 0.0819 (12.6751) | 0.0831 (18.6714) |
| SWSS-OSP | 0.1052 (7.0531) | 0.1078 (10.3613) | 0.1070 (14.7728) | 0.1074 (20.6785) |
| SWSS-N-FINDR | 0.1013 (4.7310) | 0.996 (7.8970) | 0.1023 (12.3459) | 0.1016 (18.3418) |
| SWSS-VCA | 0.0195 (4.2739) | 0.0195 (7.4921) | 0.0184 (11.9772) | 0.0193 (17.9635) |
| SWSS-AVMAX | 0.1006 (3.9692) | 0.1005 (7.1675) | 0.1020 (11.6026) | 0.1016 (17.6000) |
| Average | 0.0817 (5.0068) | 0.0818 (8.2292) | 0.0824 (12.6746) | 0.0825 (18.6460) |
Table 7.
SAD results captured from SPP coupled algorithms and improved versions under different noise and synthetic dataset scenarios.
Table 7.
SAD results captured from SPP coupled algorithms and improved versions under different noise and synthetic dataset scenarios.
| SNRs | 10 dB | 20 dB | 30 dB | 40 dB | 50 dB | 60 dB |
|---|
| SPP+OSP | 0.1859 | 0.1091 | 0.0503 | 0.0297 | 0.0178 | 0.0104 |
| SWSS-OSP | 0.4216 | 0.1527 | 0.0467 | 0.0144 | 0.0045 | 0.0014 |
| SPP+N-FINDR | 0.1618 | 0.0922 | 0.0525 | 0.0326 | 0.0184 | 0.0106 |
| SWSS-N-FINDR | 0.4004 | 0.1389 | 0.0447 | 0.0157 | 0.0045 | 0.0015 |
| SPP+VCA | 0.0858 | 0.0593 | 0.0426 | 0.0285 | 0.0173 | 0.0103 |
| SWSS-VCA | 0.0899 | 0.0205 | 0.0085 | 0.0025 | 0.0009 | 0.0003 |
| SPP+AVMAX | 0.1662 | 0.0891 | 0.0509 | 0.0299 | 0.0180 | 0.0105 |
| SWSS-AVMAX | 0.3999 | 0.1372 | 0.0447 | 0.0142 | 0.0045 | 0.0024 |
| SPP+OSP | 0.1837 | 0.1176 | 0.0531 | 0.0301 | 0.0177 | 0.0103 |
| SWSS-OSP | 0.4129 | 0.1518 | 0.0463 | 0.0142 | 0.0045 | 0.0014 |
| SPP+N-FINDR | 0.1669 | 0.0891 | 0.0554 | 0.0329 | 0.0189 | 0.0104 |
| SWSS-N-FINDR | 0.4038 | 0.1378 | 0.0445 | 0.0159 | 0.0045 | 0.0014 |
| SPP+VCA | 0.0884 | 0.0565 | 0.0424 | 0.0283 | 0.0173 | 0.0102 |
| SWSS-VCA | 0.0860 | 0.0193 | 0.0081 | 0.0026 | 0.0010 | 0.0003 |
| SPP+AVMAX | 0.1659 | 0.0919 | 0.0500 | 0.0307 | 0.0177 | 0.0103 |
| SWSS-AVMAX | 0.4023 | 0.1379 | 0.0440 | 0.0138 | 0.0044 | 0.0014 |
Table 8.
Overall results of the algorithms for experiments conducted on the dataset.
Table 8.
Overall results of the algorithms for experiments conducted on the dataset.
| Algorithms | OSP | SPP + OSP | SWSS-OSP | N-FINDR | SPP + N-FINDR | SWSS-N-FINDR | VCA | SPP + VCA | SWSS-VCA | AVMAX | SPP + AVMAX | SWSS-AVMAX |
|---|
| Alunite | 0.0859 | 0.1053 | 0.0955 | 0.0969 | 0.1086 | 0.0955 | 0.0957 | 0.1062 | 0.0944 | 0.1060 | 0.1053 | 0.0955 |
| Andradite | † | 0.0843 | 0.0990 | 0.0802 | 0.0813 | 0.0707 | 0.0691 | 0.0742 | 0.0694 | 0.0730 | 0.0713 | 0.0675 |
| Buddingtonite | 0.1159 | 0.1152 | 0.0865 | 0.0775 | 0.0921 | 0.0930 | 0.0734 | 0.1157 | 0.1216 | 0.0862 | 0.1152 | 0.1206 |
| Dumortierite | 0.0802 | 0.0681 | 0.0760 | 0.0760 | 0.0680 | 0.0760 | 0.0759 | 0.0674 | 0.0765 | 0.0780 | 0.0675 | 0.0760 |
| Kaolinite #1 | 0.0848 | 0.1047 | 0.1159 | 0.0848 | † | 0.1159 | 0.0841 | 0.0903 | † | 0.0848 | 0.0908 | 0.1159 |
| Kaolinite #2 | 0.0852 | 0.0653 | 0.0727 | 0.0695 | 0.0671 | 0.0596 | † | 0.0701 | 0.0887 | 0.0602 | 0.0699 | 0.0773 |
| Muscovite | 0.0892 | 0.0962 | 0.0892 | 0.0923 | 0.0962 | 0.0892 | 0.0914 | 0.0954 | 0.0879 | 0.0821 | 0.0912 | 0.0892 |
| Montmorillonite | 0.0618 | 0.0611 | 0.0618 | 0.0612 | 0.0600 | 0.0612 | 0.0647 | 0.0642 | 0.0628 | 0.0618 | 0.0600 | 0.0618 |
| Nontronite | 0.0736 | 0.0810 | 0.0767 | 0.0736 | 0.0810 | 0.0767 | 0.0774 | † | 0.0781 | 0.1068 | 0.0847 | 0.0767 |
| Pyrope | † | † | 0.0735 | † | † | 0.1051 | † | † | 0.0659 | † | † | 0.0671 |
| Sphene | 0.0655 | † | † | † | † | † | 0.0654 | 0.0914 | † | † | † | † |
| Chalcedony | 0.0870 | 0.1228 | 0.1328 | 0.1003 | 0.1360 | 0.1326 | 0.1144 | 0.1376 | † | 0.0933 | 0.1399 | 0.0812 |
| Average SAD | 0.0829 | 0.0904 | 0.0890 | 0.0812 | 0.0878 | 0.0887 | 0.0812 | 0.9124 | 0.0828 | 0.0832 | 0.0896 | 0.0901 |
| RMSE | 0.0335 | 0.0410 | 0.0218 | 0.0113 | 0.0347 | 0.0127 | 0.0175 | 0.0269 | 0.0108 | 0.0138 | 0.0354 | 0.0126 |
| Execution time | 39.2514 | 61.2759 | 51.9905 | 12.8841 | 61.0445 | 25.7246 | 1.1903 | 53.6188 | 11.3288 | 0.5569 | 52.8609 | 10.8062 |