Mapping Paddy Rice Planting Area in Northeastern China Using Spatiotemporal Data Fusion and Phenology-Based Method
Abstract
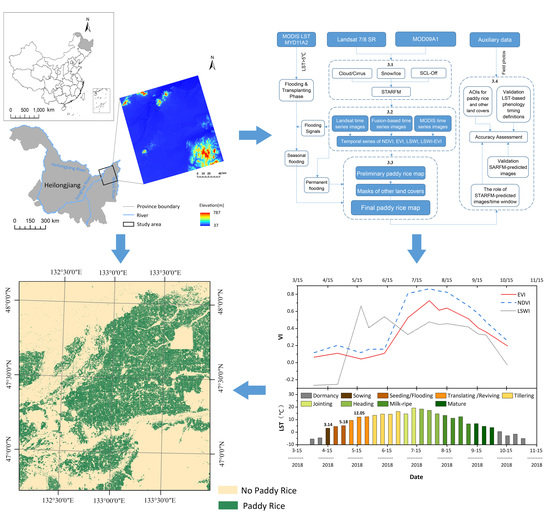
:1. Introduction
2. Study Area and Data
2.1. Study Area
2.2. Data and Pre-Processing
2.2.1. Landsat Data and Pre-Processing
2.2.2. MODIS Data and Pre-Processing
2.3. Additional Datasets
3. Method
3.1. STARFM Prediction
3.2. Fused Time Series NDVI and EVI and LSWI Data
3.3. Identification of Paddy Rice Fields
3.3.1. Algorithms to Identify Inundation/Flooding Signals
3.3.2. Implementation of Phenology-Based Paddy Rice Mapping Algorithm
- Natural vegetation. Forests, grasslands and shrubs grow earlier and are greener than paddy rice, so the EVI values of natural vegetation are higher than the EVI values of paddy field before the rice plants begin to grow. We identified the pixels with maximum EVI value ≥ 0.30 before the mid-flooding/transplanting period (corresponding to the images before May 29 in the study) as natural vegetation and generated a preliminary mask of natural vegetation. Then we combined the preliminary mask with the ALOS/PALSAR-based fine resolution (25 m) forest map (2017) to get a final natural vegetation mask.
- Sloping land. Rice plants grow in water and therefore cannot be planted in sloping land. The sloping land mask was generated based on the rule of slope ≥ 3° to exclude the areas with low probabilities of growing paddy rice by using the SRTM DEM data.
- Sparse vegetation. There are some low-vegetated lands in the field roads, built-up land, water edge and other areas. Those low-vegetated lands have very low greenness within the entire growing season (Figure 9 and Figure 11). Thus, pixels with the maximum EVI value ≤ 0.60 within the plant growing season (nighttime LST > 0 °C) were labeled as sparse vegetation. In addition, the preliminary paddy rice map had obvious strips. That was mainly because the EVI values of paddy rice pixels reached their maximum from July to August, while the images in July all contained SLC-off gaps from Landsat ETM+. Therefore, the maximum EVI values of paddy rice pixels in the strip region were mostly replaced by the paddy rice EVI values from images in September, and those paddy rice pixels were misclassified into sparse vegetation. Referring to the dates of the Landsat ETM+ images in the fused time series data, pixels in the strip region with the maximum EVI value ≤ 0.55 within the plant growing season (nighttime LST > 0 °C) were classified as sparse vegetation.
- Open canopies in permanent flooding areas, such as vegetation (grass, trees, shrubs) growing on the edge of water bodies. Pixels in the open canopies are a mixture of natural vegetation and water and have flooding signals. Therefore, it is necessary to distinguish open canopies in permanent flooded areas from open canopies in seasonally flooded areas. Unlike seasonal flooding areas such as rice fields, permanent flooded areas usually have flooding signals throughout the entire growing season. Therefore, if a pixel had the flooding signal for all images within the entire growing season, then it was marked as a permanent flooded canopy.
- Natural wetlands. There are natural wetlands in the Sanjiang Plain due to long-term waterlogging. When the temperature rises above 0 °C, natural wetlands start to flood due to snowmelt. Therefore, wetland vegetation has grown a few weeks before the flooding signals appear in paddy rice fields. The difference in EVI values between the wetland vegetation and paddy rice reaches to the maximum around the middle of June (Figure 9 and Figure 10). Therefore, if a pixel with flooding signals had maximum EVI value ≥ 0.30 between LST > 0 °C and mid-June after excluding the masks described above, then it was classified as natural wetlands.
3.4. Evaluation of Fusion-and Phenology-Based Paddy Rice Map Strategy
3.4.1. Evaluation of STARFM Prediction
3.4.2. Comparison with Other Classification Results
4. Result
4.1. Accuracy of the STARFM Predictions
4.2. Comparisons with Non-Fusion-Based and RF Classification Results
4.3. Evaluation of the Feature Importance
5. Discussion
5.1. Advantages of Fusion-and Phenology-Based Paddy Rice Mapping Strategy
5.2. Limitation and Future Opportunities
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Kuang, W.; Zhang, Z.; Xu, X.; Qin, Y.; Ning, J.; Zhou, W.; Zhang, S.; Li, R.; Yan, C.; et al. Spatiotemporal characteristics, patterns, and causes of land-use changes in China since the late 1980s. J. Geogr. Sci. 2014, 24, 195–210. [Google Scholar] [CrossRef]
- Wang, Z.B.; Hao, H.W.; Yin, X.C.; Liu, Q.; Huang, K. Exchange rate prediction with non-numerical information. Neural Comput. Appl. 2011, 20, 945–954. [Google Scholar] [CrossRef]
- Qin, Y.; Xiao, X.; Dong, J.; Zhou, Y.; Zhu, Z.; Zhang, G.; Du, G.; Jin, C.; Kou, W.; Wang, J.; et al. Mapping paddy rice planting area in cold temperate climate region through analysis of time series Landsat 8 (OLI), Landsat 7 (ETM+) and MODIS imagery. ISPRS J. Photogramm. Remote Sens. 2015, 105, 220–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, F.; Hayashi, Y.; Zhang, Z.; Sakamoto, T.; Yokozawa, M. Global warming, rice production, and water use in China: Developing a probabilistic assessment. Agric. For. Meteorol. 2008, 148, 94–110. [Google Scholar] [CrossRef]
- Gilbert, M.; Xiao, X.; Pfeiffer, D.U.; Epprecht, M.; Boles, S.; Czarnecki, C.; Chaitaweesub, P.; Kalpravidh, W.; Minh, P.Q.; Otte, M.J.; et al. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proc. Natl. Acad. Sci. USA 2008, 105, 4769–4774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, D.P.; Wulder, M.A.; Loveland, T.R.; Woodcock, C.E.; Allen, R.G.; Anderson, M.C.; Helder, D.; Irons, J.R.; Johnson, D.M.; Kennedy, R.; et al. Landsat-8: Science and product vision for terrestrial global change research. Remote Sens. Environ. 2014, 145, 154–172. [Google Scholar] [CrossRef] [Green Version]
- Kontgis, C.; Schneider, A.; Ozdogan, M. Mapping rice paddy extent and intensification in the Vietnamese Mekong River Delta with dense time stacks of Landsat data. Remote Sens. Environ. 2015, 169, 255–269. [Google Scholar] [CrossRef]
- Zhang, G.; Xiao, X.; Biradar, C.M.; Dong, J.; Qin, Y.; Menarguez, M.A.; Zhou, Y.; Zhang, Y.; Jin, C.; Wang, J.; et al. Spatiotemporal patterns of paddy rice croplands in China and India from 2000 to 2015. Sci. Total. Environ. 2017, 579, 82–92. [Google Scholar] [CrossRef]
- Zhou, Y.; Xiao, X.; Qin, Y.; Dong, J.; Zhang, G.; Kou, W.; Jin, C.; Wang, J.; Li, X. Mapping paddy rice planting area in rice-wetland coexistent areas through analysis of Landsat 8 OLI and MODIS images. Int. J. Appl. Earth Obs. Geoinf. 2016, 46, 1–12. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, Q.; Hollinger, D.; Aber, J.; Berrien, A.; Moore, B., III. Modeling gross primary production of an evergreen needleleaf forest using MODIS and climate data. Ecol. Appl. 2005, 15, 954–969. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Xiao, X.; Boles, S.; Frolking, S.; Li, C.; Babu, J.Y.; Salas, W.; Moore, B. Mapping paddy rice agriculture in South and Southeast Asia using multi-temporal MODIS images. Remote Sens. Environ. 2006, 100, 95–113. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, X.; Kou, W.; Qin, Y.; Zhang, G.; Li, L.; Jin, C.; Zhou, Y.; Wang, J.; Biradar, C.; et al. Tracking the dynamics of paddy rice planting area in 1986–2010 through time series Landsat images and phenology-based algorithms. Remote Sens. Environ. 2015, 160, 99–113. [Google Scholar] [CrossRef]
- Sakamoto, T.; Van Phung, C.; Kotera, A.; Nguyen, K.D.; Yokozawa, M. Analysis of rapid expansion of inland aquaculture and triple rice-cropping areas in a coastal area of the Vietnamese Mekong Delta using MODIS time-series imagery. Landsc. Urban Plan. 2009, 92, 34–46. [Google Scholar] [CrossRef]
- Xiao, X.; Boles, S.; Liu, J.; Zhuang, D.; Frolking, S.; Li, C.; Salas, W.; Moore, B. Mapping paddy rice agriculture in southern China using multi-temporal MODIS images. Remote Sens. Environ. 2005, 95, 480–492. [Google Scholar] [CrossRef]
- Zhang, G.; Xiao, X.; Dong, J.; Kou, W.; Jin, C.; Qin, Y.; Zhou, Y.; Wang, J.; Angelo Menarguez, M.; Biradar, C. Mapping paddy rice planting areas through time series analysis of MODIS land surface temperature and vegetation index data. ISPRS J. Photogramm. Remote Sens. 2015, 106. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, X. Evolution of regional to global paddy rice mapping methods: A review. ISPRS J. Photogramm. Remote Sens. 2016, 119, 214–227. [Google Scholar] [CrossRef] [Green Version]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Linke, J.; McDermid, G.; Masek, J.G.; Gao, F.; White, J.C. A new data fusion model for high spatial- and temporal-resolution mapping of forest disturbance based on Landsat and MODIS. Remote Sens. Environ. 2009, 113, 1613–1627. [Google Scholar] [CrossRef]
- Zhang, W.; Li, A.; Jin, H.; Bian, J.; Zhang, Z.; Lei, G.; Qin, Z.; Huang, C. An Enhanced Spatial and Temporal Data Fusion Model for Fusing Landsat and MODIS Surface Reflectance to Generate High Temporal Landsat-Like Data. Remote Sens. 2013, 5, 5346–5368. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Chen, J.; Gao, F.; Chen, X.; Masek, J.G. An enhanced spatial and temporal adaptive reflectance fusion model for complex heterogeneous regions. Remote Sens. Environ. 2010, 114, 2610–2623. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the Blending of the Landsat and MODIS Surface Reflectance: Predicting Daily Landsat Surface Reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar] [CrossRef]
- Zhu, L.; Radeloff, V.C.; Ives, A.R. Improving the mapping of crop types in the Midwestern U.S. by fusing Landsat and MODIS satellite data. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 1–11. [Google Scholar] [CrossRef]
- Wang, C.; Fan, Q.; Li, Q.; SooHoo, W.M.; Lu, L. Energy crop mapping with enhanced TM/MODIS time series in the BCAP agricultural lands. ISPRS J. Photogramm. Remote Sens. 2017, 124, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Lin, H. Object-based rice mapping using time-series and phenological data. Adv. Space Res. 2019, 63, 190–202. [Google Scholar] [CrossRef]
- Chen, B.; Huang, B.; Xu, B. Multi-source remotely sensed data fusion for improving land cover classification. ISPRS J. Photogramm. Remote Sens. 2017, 124, 27–39. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Zhang, N.; Wei, X.; Gu, X.; Zhao, X.; Yao, Y.; Xie, X. Land cover classification of finer resolution remote sensing data integrating temporal features from time series coarser resolution data. ISPRS J. Photogramm. Remote Sens. 2014, 93, 49–55. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Zhang, B.; Lu, L. Object-Based Crop Classification with Landsat-MODIS Enhanced Time-Series Data. Remote Sens. 2015, 7, 16091–16107. [Google Scholar] [CrossRef] [Green Version]
- Guan, K.; Li, Z.; Rao, L.N.; Gao, F.; Xie, D.; Hien, N.T.; Zeng, Z. Mapping Paddy Rice Area and Yields Over Thai Binh Province in Viet Nam From MODIS, Landsat, and ALOS-2/PALSAR-2. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 2238–2252. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, M.; Wu, B. Using high spatial and temporal resolution data blended from SPOT-5 and MODIS to map biomass of summer maize. In Proceedings of the 2016 Fifth International Conference on Agro-Geoinformatics (Agro-Geoinformatics), Tianjin, China, 18–20 July 2016; pp. 1–5. [Google Scholar]
- Dong, J.; Xiao, X.; Menarguez, M.A.; Zhang, G.; Qin, Y.; Thau, D.; Biradar, C.; Moore, B. Mapping paddy rice planting area in northeastern Asia with Landsat 8 images, phenology-based algorithm and Google Earth Engine. Remote Sens. Environ. 2016, 185, 142–154. [Google Scholar] [CrossRef] [Green Version]
- Senf, C.; Leitão, P.J.; Pflugmacher, D.; van der Linden, S.; Hostert, P. Mapping land cover in complex Mediterranean landscapes using Landsat: Improved classification accuracies from integrating multi-seasonal and synthetic imagery. Remote Sens. Environ. 2015, 156, 527–536. [Google Scholar] [CrossRef]
- Zhang, S.; Na, X.; Kong, B.; Wang, Z.; Jiang, H.; Yu, H.; Zhao, Z.; Li, X.; Liu, C.; Dale, P. Identifying Wetland Change in China’s Sanjiang Plain Using Remote Sensing. Wetlands 2009, 29. [Google Scholar] [CrossRef]
- Lu, Z.J.; Qian, S.O.; Liu, K.B.; Wu, W.B.; Liu, Y.X.; Rui, X.I.; Zhang, D.M. Rice cultivation changes and its relationships with geographical factors in Heilongjiang Province, China. J. Integr. Agric. 2017, 16, 2274–2282. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wan, W.; Fang, Y.; Zhu, S.; Chen, X.; Liu, B.; Yang, H. A Google Earth Engine-enabled Software for Efficiently Generating High-quality User-ready Landsat Mosaic Images. Environ. Model. Softw. 2018, 112. [Google Scholar] [CrossRef]
- Masek, J.; Vermote, E.; Saleous, N.; Wolfe, R.; Hall, F.; Huemmrich, K.F.; Gao, F.; Kutler, J.; Lim, T.-K. A Landsat Surface Reflectance Data Set for North America, 1990–2000. IEEE Geosci. Remote Sens. Lett. 2006, 3, 68–72. [Google Scholar] [CrossRef]
- Vermote, E.; Justice, C.; Claverie, M.; Franch, B. Preliminary analysis of the performance of the Landsat 8/OLI land surface reflectance product. Remote Sens. Environ. 2016, 185, 46–56. [Google Scholar] [CrossRef]
- Zhu, Z.; Woodcock, C.E. Object-based cloud and cloud shadow detection in Landsat imagery. Remote Sens. Environ. 2012, 118, 83–94. [Google Scholar] [CrossRef]
- Shimada, M.; Itoh, T.; Motooka, T.; Watanabe, M.; Shiraishi, T.; Thapa, R.; Lucas, R. New global forest/non-forest maps from ALOS PALSAR data (2007–2010). Remote Sens. Environ. 2014, 155, 13–31. [Google Scholar] [CrossRef]
- Cui, J.; Zhang, X.; Luo, M. Combining Linear Pixel Unmixing and STARFM for Spatiotemporal Fusion of Gaofen-1 Wide Field of View Imagery and MODIS Imagery. Remote Sens. 2018, 10, 1047. [Google Scholar] [CrossRef]
- Yang, G.; Weng, Q.; Pu, R.; Gao, F.; Sun, C.; Li, H.; Zhao, C. Evaluation of ASTER-Like Daily Land Surface Temperature by Fusing ASTER and MODIS Data during the HiWATER-MUSOEXE. Remote Sens. 2016, 8, 75. [Google Scholar] [CrossRef]
- Xiao, X.; Boles, S.; Frolking, S.; Salas, W.; Moore, B.; Li, C.; He, L.; Zhao, R. Observation of flooding and rice transplanting of paddy rice fields at the site to landscape scales in China using VEGETATION sensor data. Int. J. Remote Sens. 2002, 23, 3009–3022. [Google Scholar] [CrossRef]
- Xiao, X.; Braswell, B.; Zhang, Q.; Boles, S.; Frolking, S.; Moore, B. Sensitivity of vegetation indices to atmospheric aerosols: Continental-scale observations in Northern Asia. Remote Sens. Environ. 2003, 84, 385–392. [Google Scholar] [CrossRef]
- Cheng, Q. Multisensor Comparisons for Validation of MODIS Vegetation Indices1 1Project supported by the National Natural Science Foundation of China (No. 40171065) and the National High Technology Research and Development Program (863 Program) of China (No. 2002AA243011). Pedosphere 2006, 16, 362–370. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chang, Q.; Li, F.; Yan, L.; Huang, Y.; Wang, Q.; Luo, L. Effects of Growth Stage Development on Paddy Rice Leaf Area Index Prediction Models. Remote Sens. 2019, 11, 361. [Google Scholar] [CrossRef]
- Ke, Y.; Im, J.; Park, S.; Gong, H. Spatiotemporal downscaling approaches for monitoring 8-day 30 m actual evapotranspiration. ISPRS J. Photogramm. Remote Sens. 2017, 126, 79–93. [Google Scholar] [CrossRef]
- Gevaert, C.M.; García-Haro, F.J. A comparison of STARFM and an unmixing-based algorithm for Landsat and MODIS data fusion. Remote Sens. Environ. 2015, 156, 34–44. [Google Scholar] [CrossRef]















| Prediction Test | Input Landsat | Input MODIS | Input MODIS | Validation Landsat |
|---|---|---|---|---|
| 1 | 19/05/2018 | 17/05/2018 | 25/05/2018 | 27/05/2018 |
| 2 | 27/05/2018 | 25/05/2018 | 17/05/2018 | 19/05/2018 |
| 3 | 06/07/2018 | 04/07/2018 | 05/08/2018 | 07/08/2018 |
| Band | Prediction Test 1 | Prediction Test 2 | Prediction Test 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Blue | 0.02 | 0.80 | 0.01 | 0.02 | 0.79 | 0.01 | 0.01 | 0.68 | 0.01 |
| Green | 0.02 | 0.83 | 0.01 | 0.02 | 0.82 | 0.01 | 0.01 | 0.72 | 0.01 |
| Red | 0.02 | 0.89 | 0.02 | 0.02 | 0.88 | 0.02 | 0.02 | 0.74 | 0.01 |
| NIR | 0.04 | 0.93 | 0.03 | 0.04 | 0.91 | 0.03 | 0.08 | 0.67 | 0.05 |
| SWIR1 | 0.04 | 0.92 | 0.03 | 0.04 | 0.92 | 0.03 | 0.04 | 0.73 | 0.03 |
| SWIR2 | 0.04 | 0.91 | 0.03 | 0.04 | 0.92 | 0.03 | 0.04 | 0.66 | 0.02 |
| VI | Prediction Test 1 | Prediction Test 2 | Prediction Test 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NDVI | 0.13 | 0.90 | 0.07 | 0.09 | 0.94 | 0.06 | 0.13 | 0.95 | 0.05 |
| EVI | 0.05 | 0.71 | 0.03 | 0.05 | 0.96 | 0.03 | 0.27 | 0.96 | 0.16 |
| LSWI | 0.20 | 0.86 | 0.11 | 0.15 | 0.89 | 0.09 | 0.14 | 0.82 | 0.06 |
| Paddy Rice Map | Class | PA (%) | UA (%) | OA (%) | Kappa Coefficient |
|---|---|---|---|---|---|
| Fusion-based | Paddy rice | 97.96 | 98.42 | 98.19 | 0.96 |
| Others | 98.42 | 97.95 | |||
| MODIS-based | Paddy rice | 82.84 | 95.09 | 89.23 | 0.78 |
| Others | 95.68 | 84.68 | |||
| Landsat-based | Paddy rice | 90.91 | 93.23 | 92.12 | 0.84 |
| Others | 93.34 | 91.05 | |||
| RF-based | Paddy rice | 96.20 | 91.38 | 93.53 | 0.87 |
| Others | 90.85 | 95.95 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Q.; Liu, M.; Cheng, J.; Ke, Y.; Chen, X. Mapping Paddy Rice Planting Area in Northeastern China Using Spatiotemporal Data Fusion and Phenology-Based Method. Remote Sens. 2019, 11, 1699. https://doi.org/10.3390/rs11141699
Yin Q, Liu M, Cheng J, Ke Y, Chen X. Mapping Paddy Rice Planting Area in Northeastern China Using Spatiotemporal Data Fusion and Phenology-Based Method. Remote Sensing. 2019; 11(14):1699. https://doi.org/10.3390/rs11141699
Chicago/Turabian StyleYin, Qi, Maolin Liu, Junyi Cheng, Yinghai Ke, and Xiuwan Chen. 2019. "Mapping Paddy Rice Planting Area in Northeastern China Using Spatiotemporal Data Fusion and Phenology-Based Method" Remote Sensing 11, no. 14: 1699. https://doi.org/10.3390/rs11141699