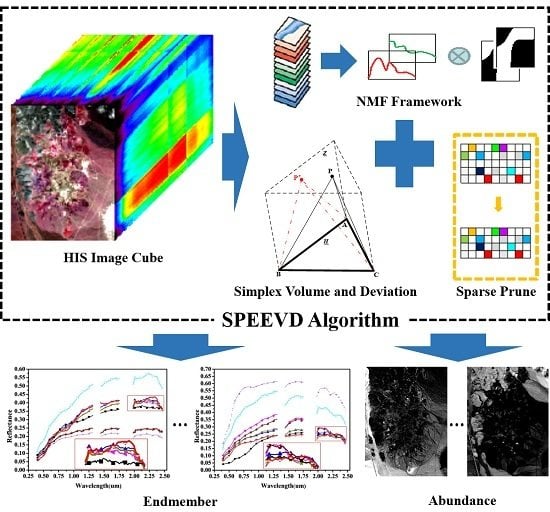
An Automatic Sparse Pruning Endmember Extraction Algorithm with a Combined Minimum Volume and Deviation Constraint
Abstract
:1. Introduction
2. Review of the ICE Algorithm
3. The SPEEVD Algorithm
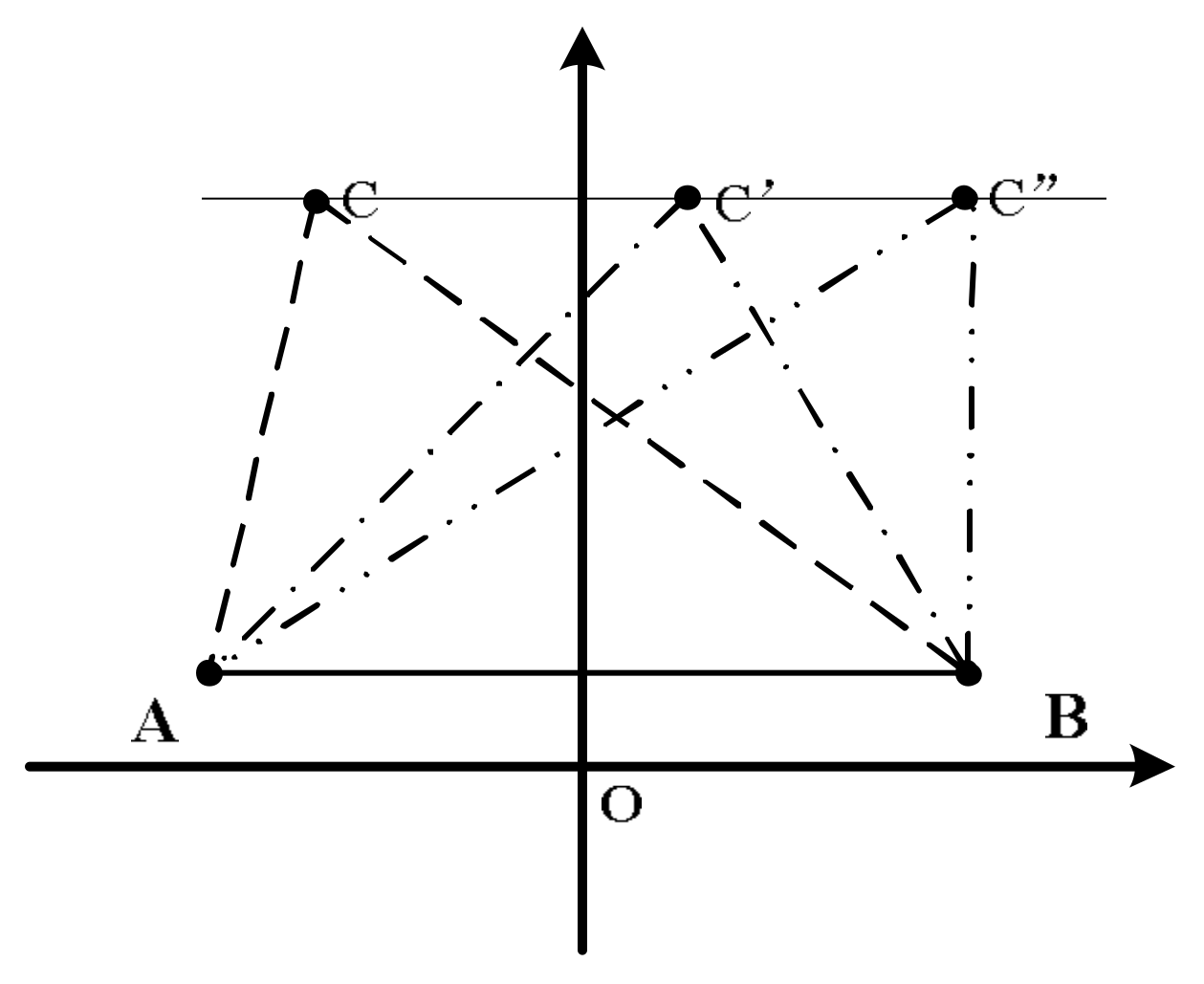
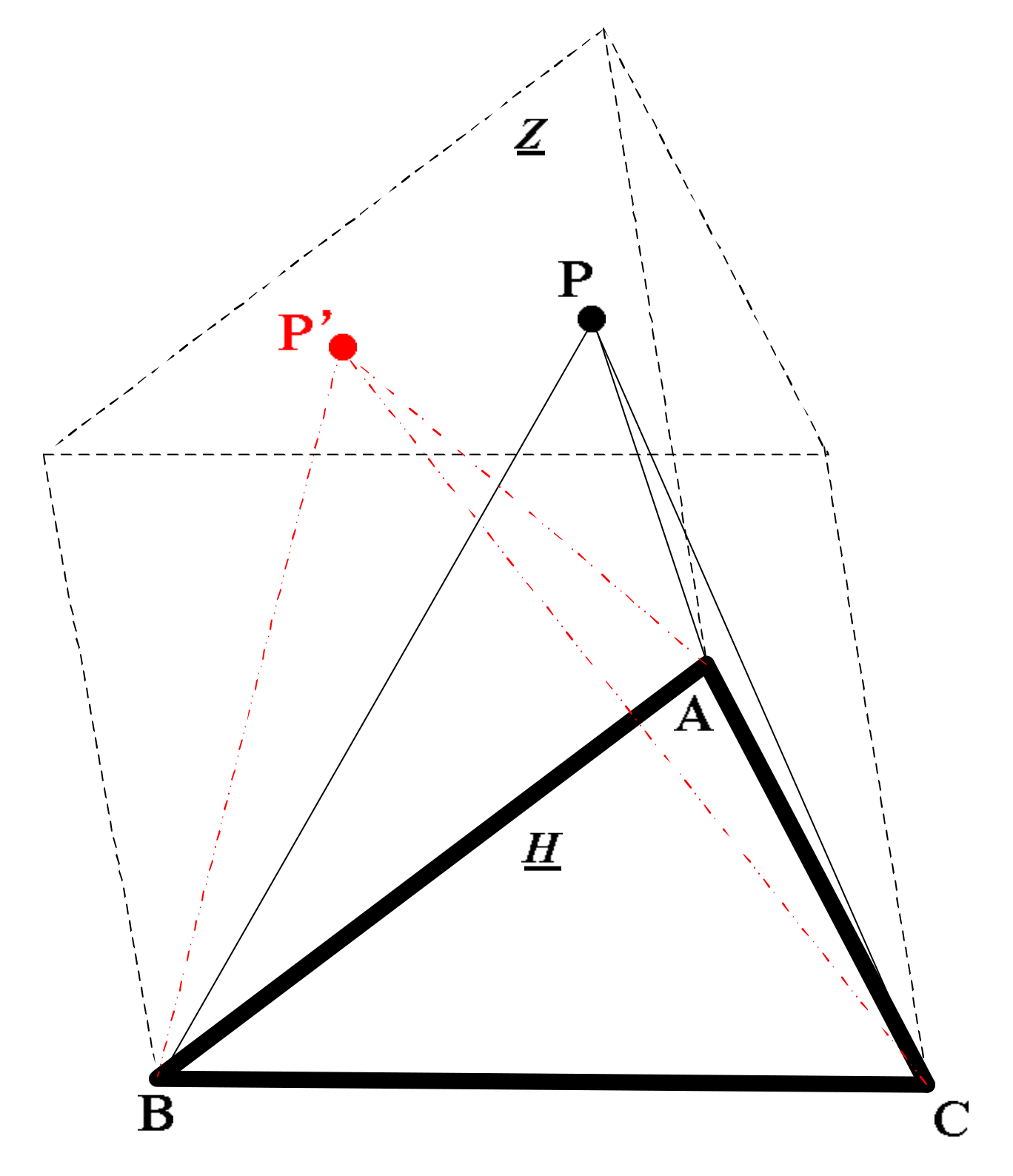
3.1. Volume and Deviation Constraint By Cross Product Between Vectors
3.2. Sparse Pruning
3.3. SPEEVD
3.3.1. Problem Formulation
3.3.2. Pre-Processing and Initialization
3.3.3. Optimization Algorithm Description
3.3.4. Stopping Criterions
| Algorithm 1: The procedure of the SPEEVD framework. |
| Input: hyperspectral image data |
| (1) Pre-processing: Use the first M bands data after MNF transformation as observation data, where the value of M is obtained according to Figure 3. (2) Initialization: k = 0. Choose the regularization parameter , parameter T, threshold Tr, endmember matrix E. (3) Compute D using the formulation (11), and combine with E, using a quadratic program to solve P. Update , while k←k + 1 (4) Then, with the latest P, renew in the E set by the formulation (28). Prune the endmember vector from the E matrix whose corresponding abundance is below the threshold Tp, as Equation (19): , where D is a scale, using the latest iteration results of E to compute by , ensuring that E is non-negative, which is physically significant. Update While k←k + 1 (5) Repeat this cycle (3)–(4) until the stopping criterion is satisfied. (6) Execute inverse MNF transform to E, obtain Ê. |
| Output: endmember matrix Ê and abundance matrix P. |
4. Experiments and Analysis
4.1. Experiment with Simulated Images
4.1.1. Experiment 1
4.1.2. Experiment 2
4.1.3. Experiment 3
4.1.4. Experiment 4
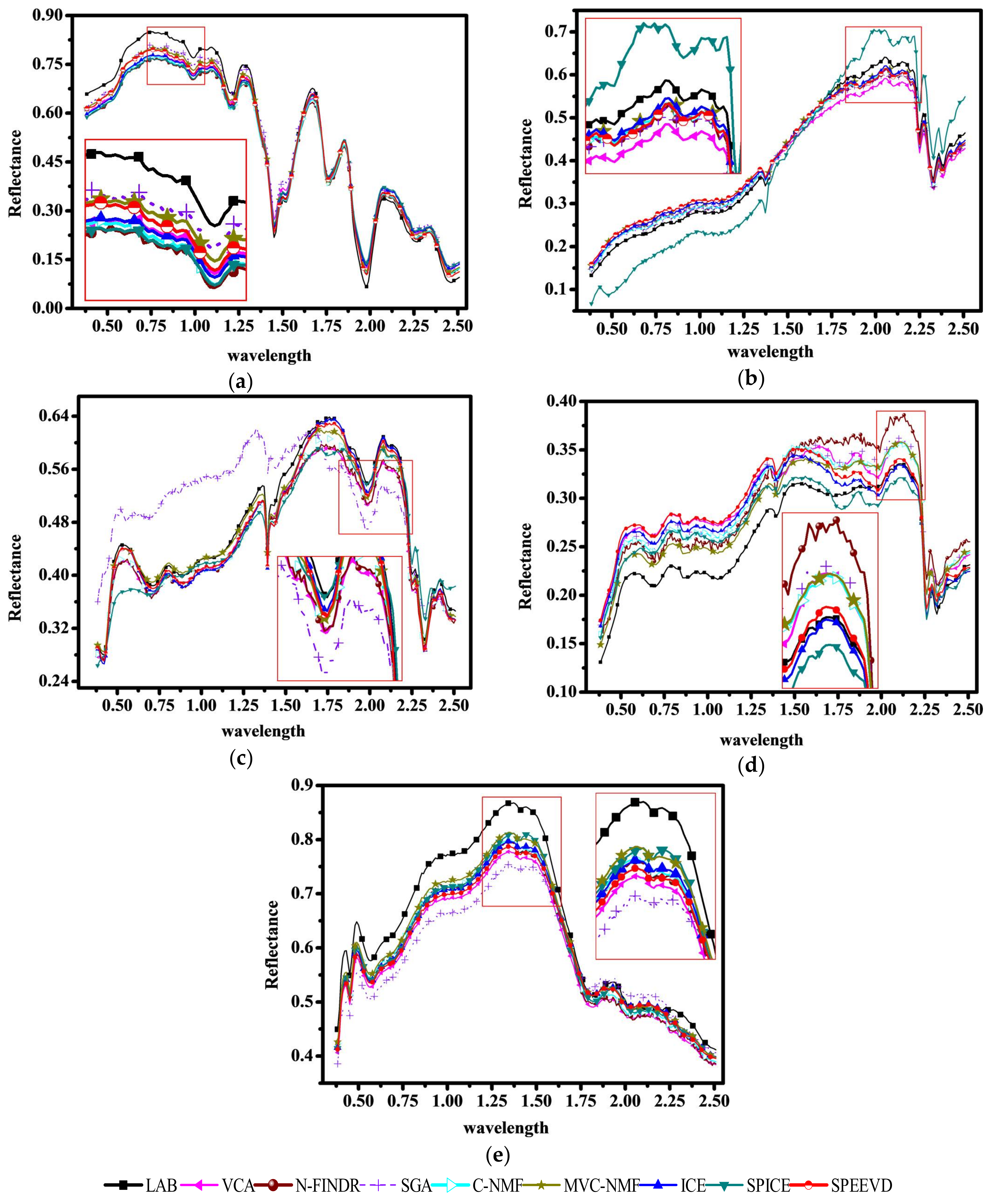
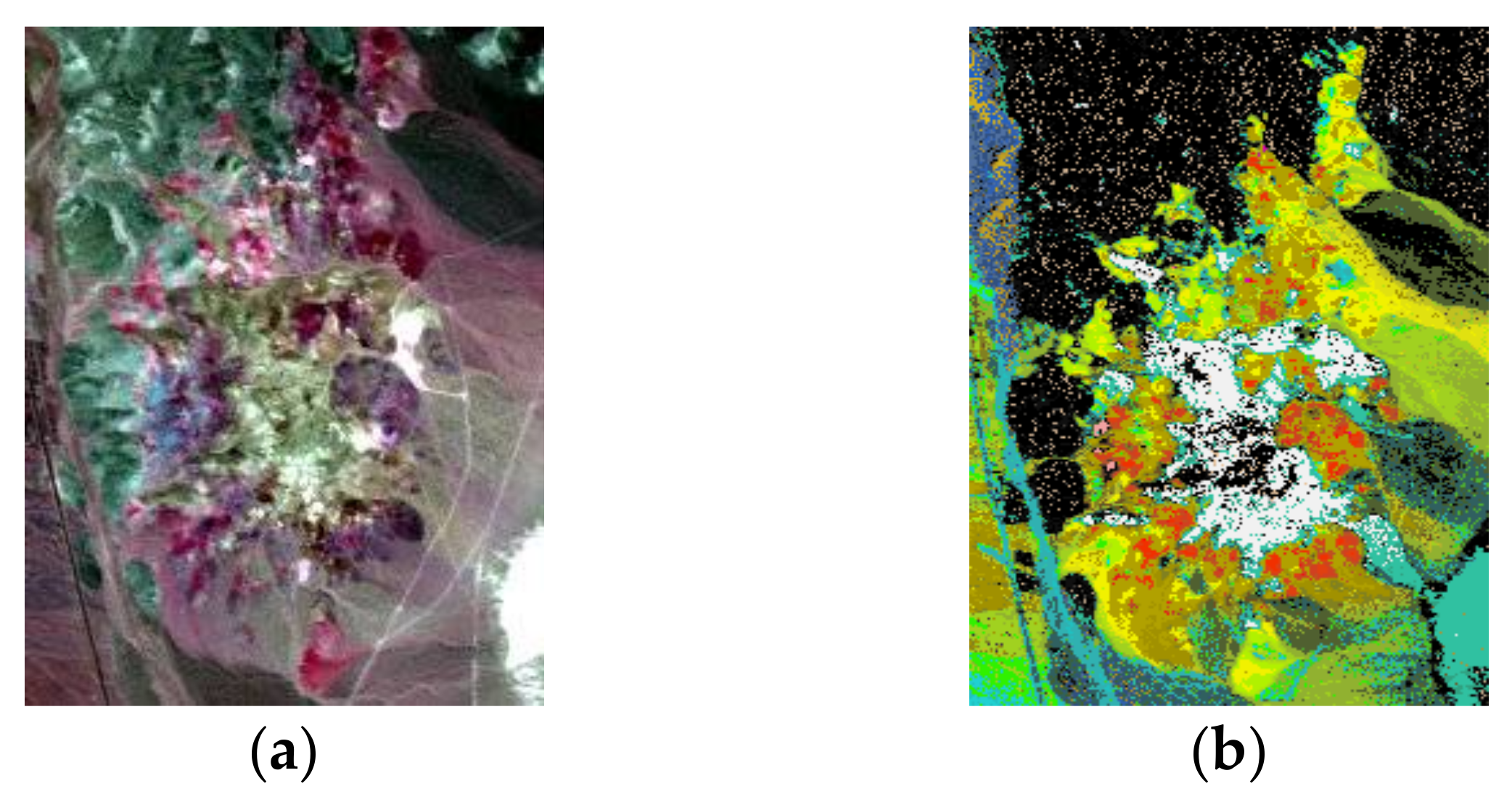
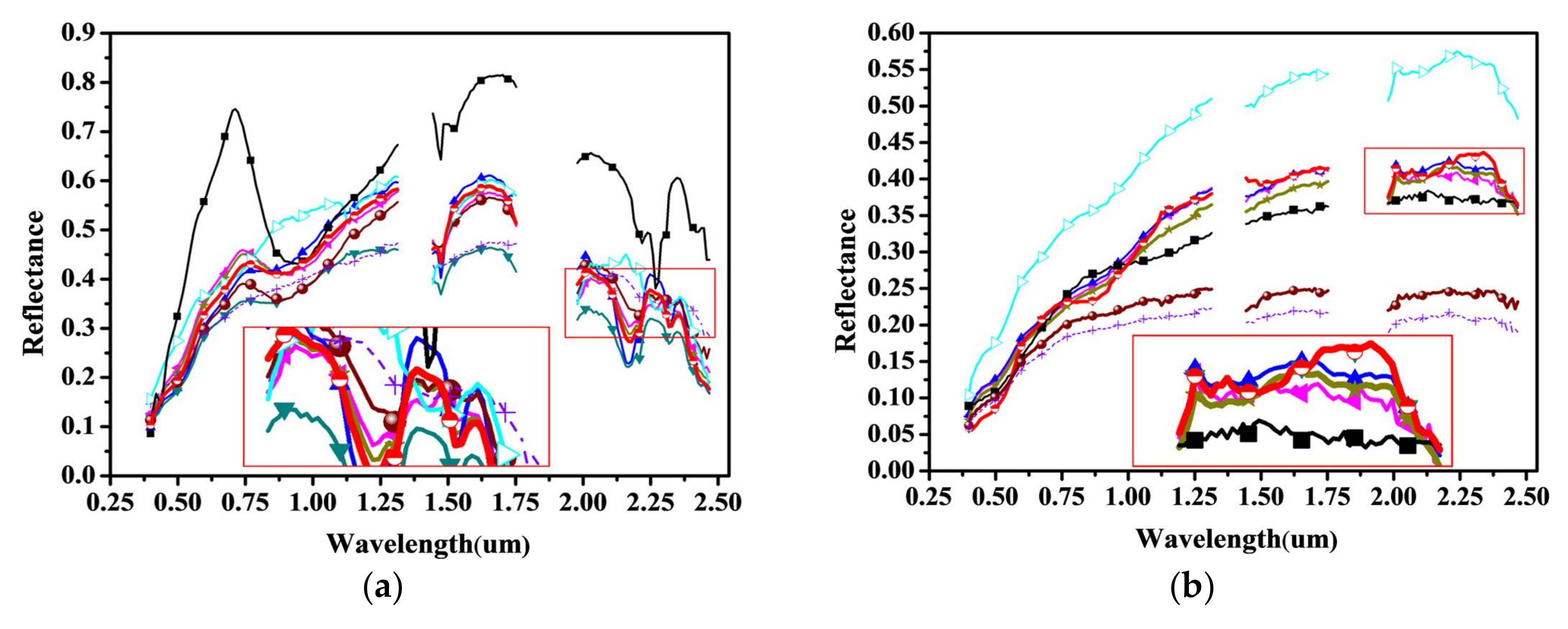
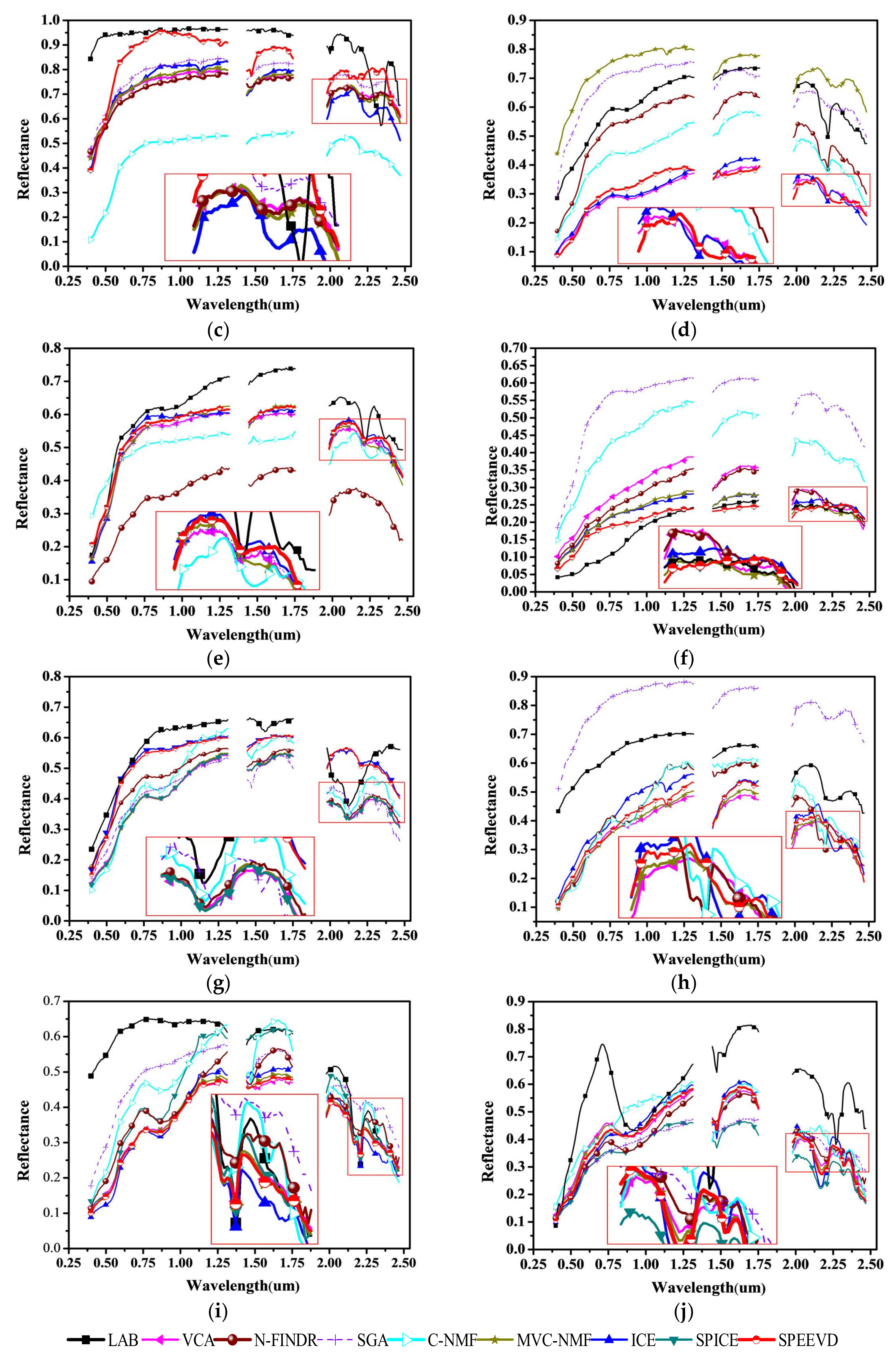
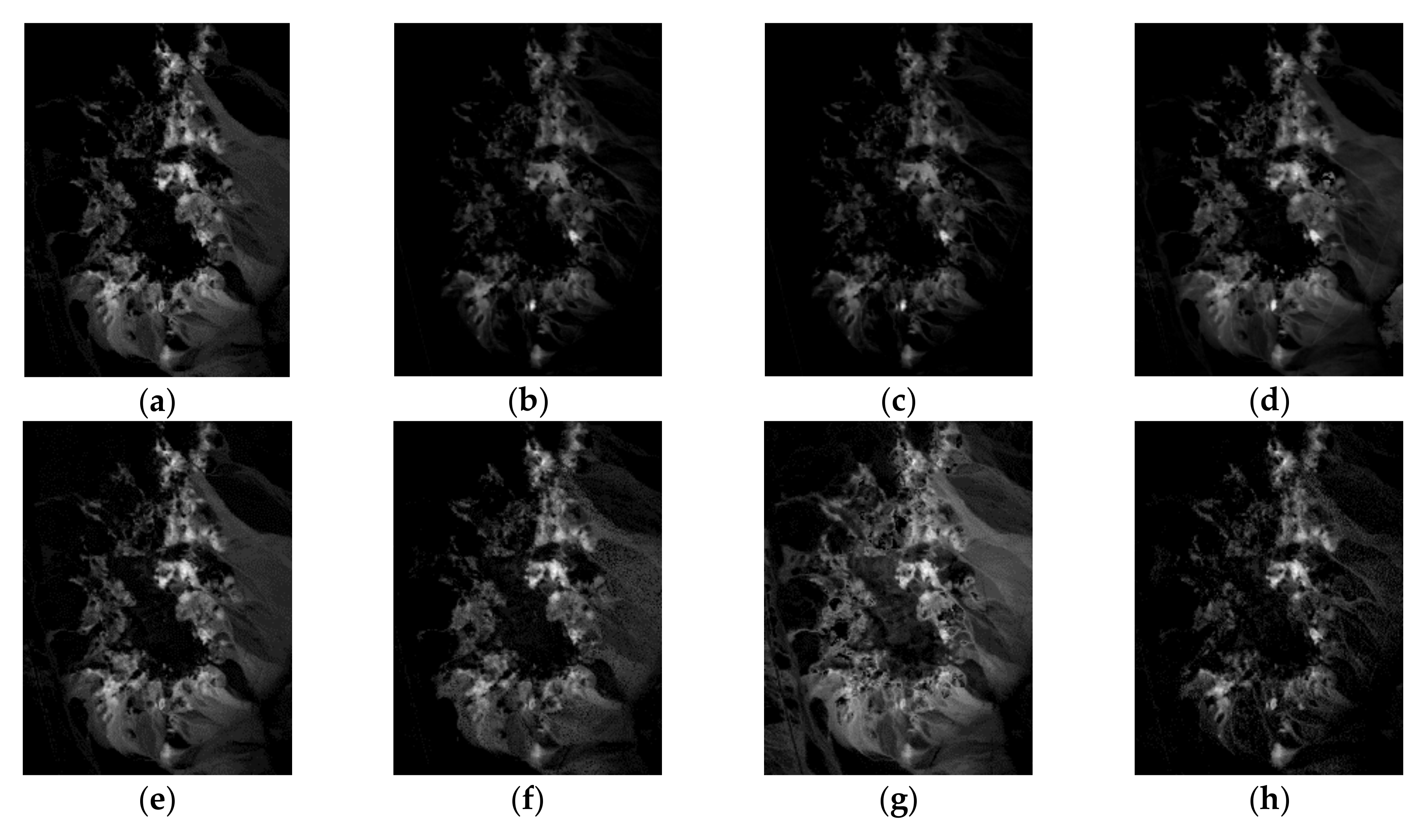
4.2. Experiments with Cuprite Image AVIRIS Data
4.3. Experiments with AVIRIS Lunar Crater Volcanic Field Data
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pan, L.; Li, H.C.; Deng, Y.J.; Zhang, F.; Chen, X.D.; Du, Q. Hyperspectral Dimensionality Reduction by Tensor Sparse and Low-Rank Graph-Based Discriminant Analysis. Remote Sens. 2017, 9, 452. [Google Scholar] [CrossRef]
- Feng, F.B.; Li, W.; Du, Q.; Zhang, B. Dimensionality Reduction of Hyperspectral Image with Graph-Based Discriminant Analysis Considering Spectral Similarity. Remote Sens. 2017, 9, 323. [Google Scholar] [CrossRef]
- Zare, A.; Gader, P. Hyperspectral Band Selection and Endmember Detection Using Sparsity Promoting Priors. IEEE Geosci. Remote Sens. Lett. 2008, 5, 256–260. [Google Scholar] [CrossRef]
- Renard, N.; Bourennane, S. Dimensionality Reduction Based on Tensor Modeling for Classification Methods. IEEE Trans. Geosci. Remote Sens. 2009, 47, 1123–1131. [Google Scholar] [CrossRef]
- Garcia, R.A.; Lee, Z.P.; Hochberg, E.J. Hyperspectral Shallow-Water Remote Sensing with an Enhanced Benthic Classifier. Remote Sens. 2018, 10, 147. [Google Scholar] [CrossRef]
- Du, B.; Zhang, L. Target detection based on a dynamic subspace. Pattern Recogn. 2014, 47, 344–358. [Google Scholar] [CrossRef]
- Du, B.; Zhang, L. Random Selection Based Anomaly Detector for Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2011, 49, 1578–1589. [Google Scholar] [CrossRef]
- Du, B.; Zhang, L.P. A discriminative metric learning based anomaly detection method. IEEE Trans. Geosci. Remote Sens. 2014, 52, 6844–6857. [Google Scholar]
- Qian, S. Hyperspectral data compression using a fast vector quantization algorithm. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1791–1798. [Google Scholar] [CrossRef]
- Du, Q.; Zhu, W.; Fowler, J.E. Anomaly-Based JPEG2000 Compression of Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2008, 5, 696–700. [Google Scholar] [CrossRef]
- Lanaras, C.; Baltsavias, E.; Schindler, K. Hyperspectral Super-Resolution with Spectral Unmixing Constraints. Remote Sens. 2017, 9, 1196. [Google Scholar] [CrossRef]
- Rizkinia, M.; Okuda, M. Joint Local Abundance Sparse Unmixing for Hyperspectral Images. Remote Sens. 2017, 9, 1224. [Google Scholar] [CrossRef]
- Feng, R.Y.; Zhong, Y.F.; Wang, L.Z.; Lin, W.J. Rolling Guidance Based Scale-Aware Spatial Sparse Unmixing for Hyperspectral Remote Sensing Imagery. Remote Sens. 2017, 9, 1218. [Google Scholar] [CrossRef]
- Iordache, M.D.; Bioucas-Dias, J.M.; Plaza, A. Sparse Unmixing of Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2014–2039. [Google Scholar] [CrossRef]
- Greer, J.B. Sparse Demixing of Hyperspectral Images. IEEE Trans. Image Process. 2012, 21, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Xiong, W.; Wu, J.; Zhang, L.; Zhang, L.; Tao, D. Stacked Convolutional Denoising Auto-Encoders for Feature Representation. IEEE Trans. Cybern. 2017, 47, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Iordache, M.-D.; Bioucas-Dias, J.M.; Plaza, A. Total Variation Spatial Regularization for Sparse Hyperspectral Unmixing. IEEE Trans. Geosci. Remote Sens. 2012, 50, 4484–4502. [Google Scholar] [CrossRef]
- Themelis, K.E.; Rontogiannis, A.A.; Koutroumbas, K.D. A Novel Hierarchical Bayesian Approach for Sparse Semisupervised Hyperspectral Unmixing. IEEE Trans. Signal Process. 2012, 60, 585–599. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Jia, X.; Somers, B.; Wu, J.; Coppin, P. A Quantitative Analysis of Virtual Endmembers' Increased Impact on the Collinearity Effect in Spectral Unmixing. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2945–2956. [Google Scholar] [CrossRef]
- Plaza, J.; Plaza, A.; Martinez, P.; Perez, R. H-COMP: A tool for quantitative and comparative analysis of endmember identification algorithms. Proc. Geosci. Remote Sens. Symp. 2003, 1, 291–293. [Google Scholar]
- Boardman, J.; Kruse, F.; Green, R. Mapping target signatures via partial unmixing of AVIRIS data. In Proceedings of the Summaries JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 23–26 January 1995; pp. 23–26. [Google Scholar]
- Du, B.; Zhang, M.; Zhang, L.; Hu, R.; Tao, D. PLTD: Patch-Based Low-Rank Tensor Decomposition for Hyperspectral Images. IEEE Trans. Multimedia 2017, 19, 67–79. [Google Scholar] [CrossRef]
- Du, B.; Zhang, Y.; Zhang, L.; Tao, D. Beyond the Sparsity-Based Target Detector: A Hybrid Sparsity and Statistics Based Detector for Hyperspectral Images. IEEE Trans. Image Process. 2016, 25, 5345–5357. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.E. N-finder: An algorithm for fast autonomous spectral endmember determination in hyperspectral data. Proc. SPIE 1999, 3753, 266–275. [Google Scholar]
- Nascimento, J.M.P.; Bioucas-Dias, J.M. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Chang, C.I.; Wu, C.C.; Liu, W.; Ouyang, Y.C. A New Growing Method for Simplex-Based Endmember Extraction Algorithm. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2804–2819. [Google Scholar] [CrossRef]
- Wu, C.C.; Lo, C.S.; Chang, C.I. Improved Process for Use of a Simplex Growing Algorithm for Endmember Extraction. IEEE Geosci. Remote Sens. Lett. 2009, 6, 523–527. [Google Scholar]
- Plaza, A.; Martinez, P.; Perez, R.; Plaza, J. Spatial/Spectral Endmember Extraction by Multidimensional Morphological Operations. IEEE Trans. Geosci. Remote Sens. 2002, 40, 756–770. [Google Scholar] [CrossRef]
- Ifarraguerri, A.; Chang, C.I. Multispectral and hyperspectral image analysis with convex cones. IEEE Trans. Geosci. Remote Sens. 1999, 37, 756–770. [Google Scholar] [CrossRef]
- Craig, M. Minimum volume transforms for remotely sensed data. IEEE Trans. Geosci. Remote Sens. 1994, 32, 542–552. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J. Minimum volume simplex analysis: A fast algorithm to unmix hyperspectral data. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Boston, MA, USA, 7–11 July 2008; pp. 2369–2371. [Google Scholar]
- Chan, T.H.; Chi, C.Y.; Huang, Y.M.; Ma, W.K. A Convex Analysis-Based Minimum-Volume Enclosing Simplex Algorithm for Hyperspectral Unmixing. IEEE Trans. Signal Process. 2009, 57, 4418–4432. [Google Scholar] [CrossRef]
- Liu, X.; Xia, W.; Wang, B.; Zhang, L. An approach based on constrained nonnegative matrix factorization to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2011, 49, 757–772. [Google Scholar] [CrossRef]
- Jia, S.; Qian, Y. Constrained nonnegative matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2009, 47, 161–173. [Google Scholar] [CrossRef]
- Parra, L.C.; Sajda, P.; Du, S. Recovery of constituent spectra using non-negative matrix factorization. Proc. SPIE 2003, 1, 321–331. [Google Scholar]
- Miao, L.; Qi, H. Endmember extraction from highly mixed data using minimum volume constrained nonnegative matrix factorization. IEEE Trans. Geosci. Remote Sens. 2007, 45, 765–777. [Google Scholar] [CrossRef]
- Bioucas-Dias, M. A variable splitting augmented Lagrangian approach to linear spectral unmixing. In Proceedings of the First Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Grenoble, France, 26–28 August 2009; pp. 1–4. [Google Scholar]
- Neville, R.A.; Staenz, K.; Szeredi, T.; Lefebvre, J.; Hauff, P. Automatic endmember extraction from hyperspectral data for mineral exploration. In Proceedings of the 21st Canadian Symposium Remote Sensing, Ottawa, ON, Canada, 21–24 June 1999; pp. 21–24. [Google Scholar]
- Li, H.; Zhang, L. A Hybrid Automatic Endmember Extraction Algorithm Based on a Local Window. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4223–4238. [Google Scholar] [CrossRef]
- Berman, M.; Kiiveri, H.; Lagerstrom, R.; Ernst, A.; Dunne, R.; Huntington, J.F. ICE: A statistical approach to identifying endmembers in hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2004, 42, 2085–2095. [Google Scholar] [CrossRef]
- Zare, A.; Gader, P. Sparsity Promoting Iterated Constrained Endmember Detection in Hyperspectral Imagery. IEEE Geosci. Remote Sens. Lett. 2007, 4, 446–450. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, G.; Xie, S.; Ding, S.; Yang, J.; Zhang, J. Blind Spectral Unmixing Based on Sparse Nonnegative Matrix Factorization. IEEE Trans. Image Process. 2011, 20, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wu, H.; Yuan, Y.; Yan, P.; Li, X. Manifold Regularized Sparse NMF for Hyperspectral Unmixing. IEEE Trans. Geosci. Remote Sens. 2012, 99, 1–12. [Google Scholar] [CrossRef]
- Chang, C.I.; Du, Q. Estimation of Number of Spectrally Distinct Signal Sources in Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 608–619. [Google Scholar] [CrossRef]
- Bajorski, P. Second Moment Linear Dimensionality as an Alternative to Virtual Dimensionality. IEEE Trans. Geosci. Remote Sens. 2011, 49, 672–678. [Google Scholar] [CrossRef]
- Luo, B.; Chanussot, J.; Douté, S. Unsupervised endmember extraction: Application to hyperspectral images from Mars. In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009. [Google Scholar]
- Bioucas-Dias, J.M.; Nascimento, J.M.P. Hyperspectral Subspace Identification. IEEE Trans. Geosci. Remote Sens. 2008, 46, 2435–2445. [Google Scholar] [CrossRef]
- Kuybeda, O.; Malah, D.; Barzohar, M. Rank estimation and redundancy reduction of high-dimensional noisy signals with preservation of rare vectors. IEEE Trans. Signal Process. 2007, 55, 5579–5592. [Google Scholar] [CrossRef]
- Acito, N.; Diani, M.; Corsini, G. Hyperspectral Signal Subspace Identification in the presence of rare signal components. IEEE Trans. Geosci. Remote Sens. 2010, 48, 1940–1954. [Google Scholar] [CrossRef]
- Fukunaga, K. Intrinsic dimensionality extraction. Classif. Pattern Recognit. Reduct. Dimens. 1982, 2, 347–360. [Google Scholar]
- Lee, D.; Seung, H.S. Algorithms for Non-Negative Matrix Factorization. Available online: https://papers.nips.cc/paper/1861-algorithms-for-non-negative-matrix-factorization.pdf (accessed on 1 March 2018).
- Hoyer, P.O. Non-negative Matrix Factorization with Sparseness Constraints. J. Mach. Learn. Res. 2004, 5, 1457–1469. [Google Scholar]
- Li, C.; Ma, Y.; Mei, X.G.; Fan, F.; Huang, J.; Ma, J.Y. Sparse Unmixing of Hyperspectral Data with Noise Level Estimation. Remote Sens. 2017, 9, 1166. [Google Scholar] [CrossRef]
- Leon, S.J. Linear Algebra with Applications, 7th ed.; China Machine Press: Beijing, China, 2009. [Google Scholar]
- Figueiredo, M.A.T. Adaptive sparseness for supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2003, 25, 1150–1159. [Google Scholar] [CrossRef]
- Casazza, P.G.; Heinecke, A.; Krahmer, F.; Kutyniok, G. Optimally Sparse Frames. IEEE Trans. Inf. Theory 2011, 99, 1–16. [Google Scholar] [CrossRef]
- Williams, P. Bayesian regularization and pruning using a Laplace prior. Neural Comput. 1995, 7, 117–143. [Google Scholar] [CrossRef]
- USGS Spectroscopy Lab. Cuprite, Nevada, AVIRIS 1995 Data. Available online: http://speclab.cr.usgs.gov/cuprite95.1um_map.tgif.gif (accessed on 1 March 2018).
- Hendrix, E.M.T.; Garcia, I.; Plaza, J.; Martin, G.; Plaza, A. A New Minimum-Volume Enclosing Algorithm for Endmember Identification and Abundance Estimation in Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2012, 50, 2744–2757. [Google Scholar] [CrossRef]

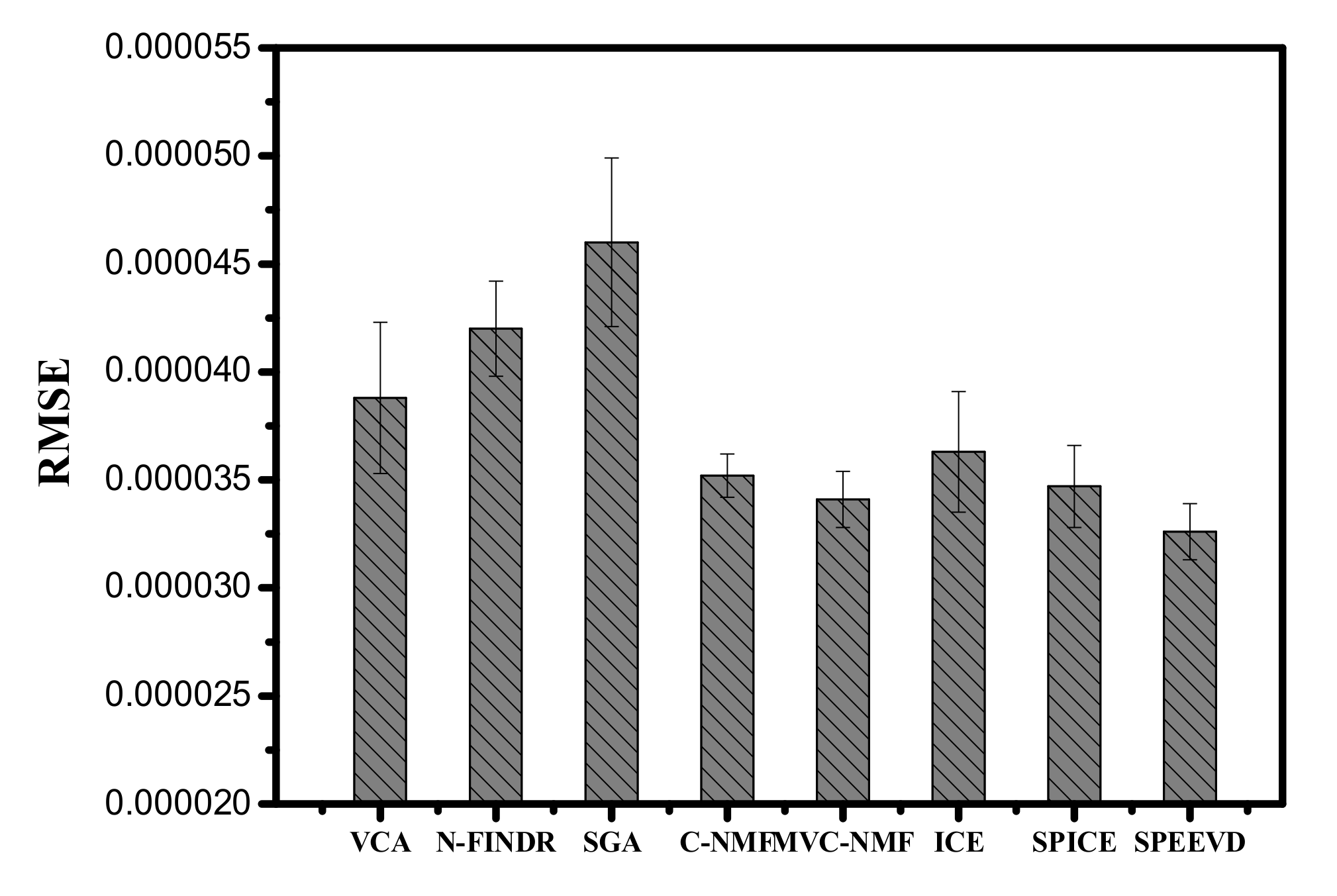
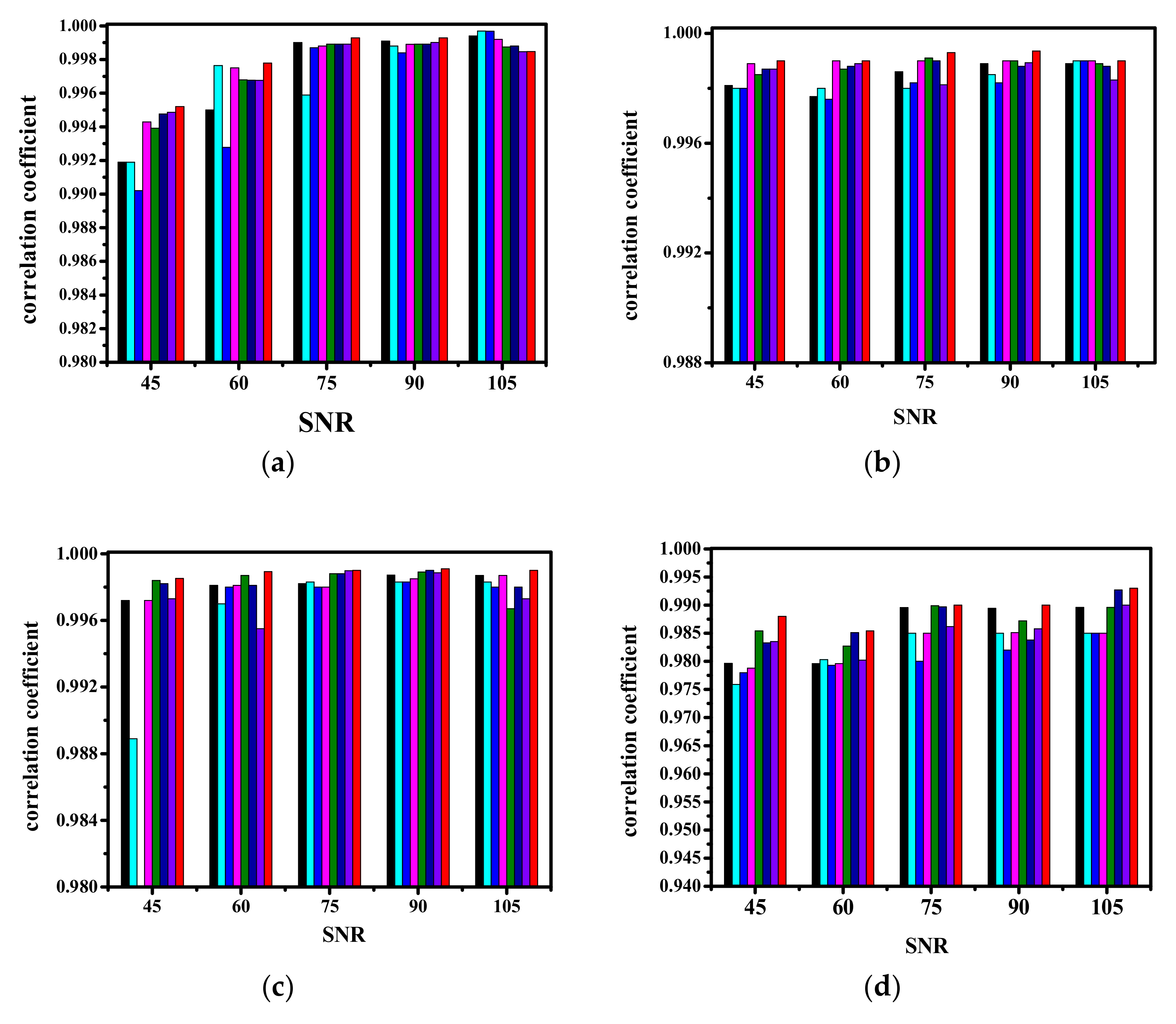

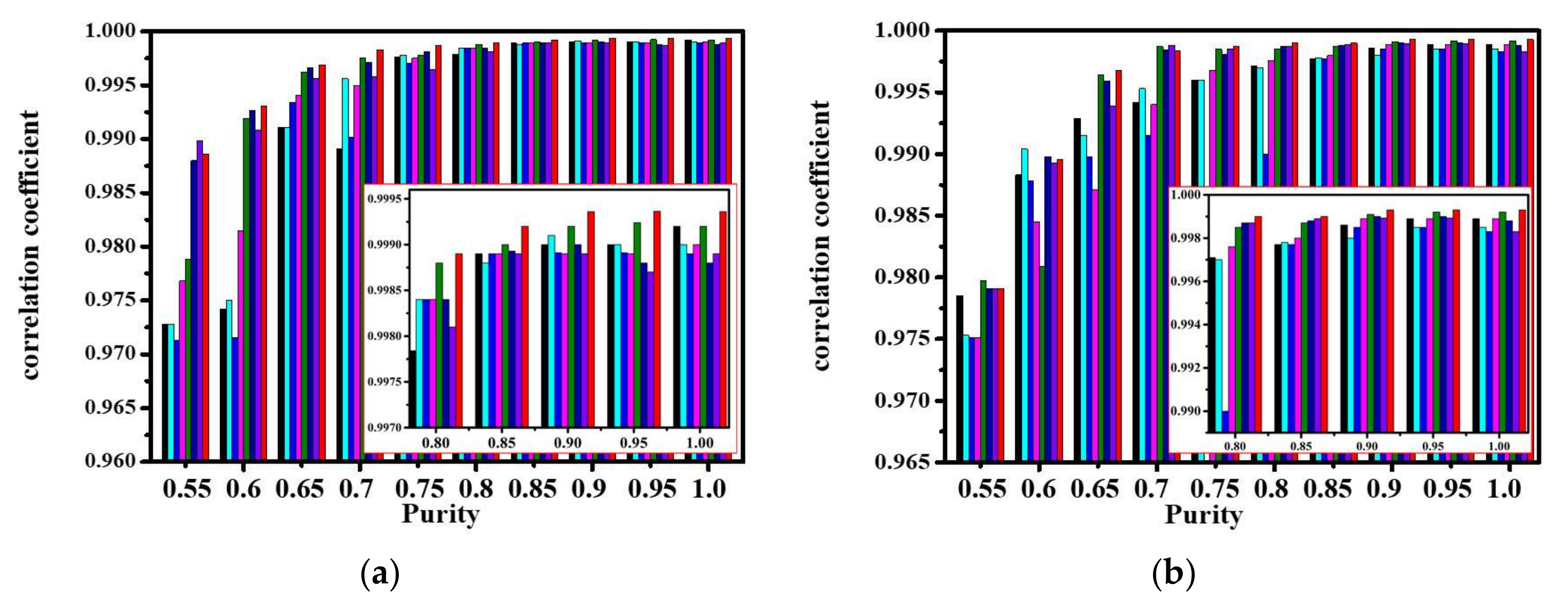
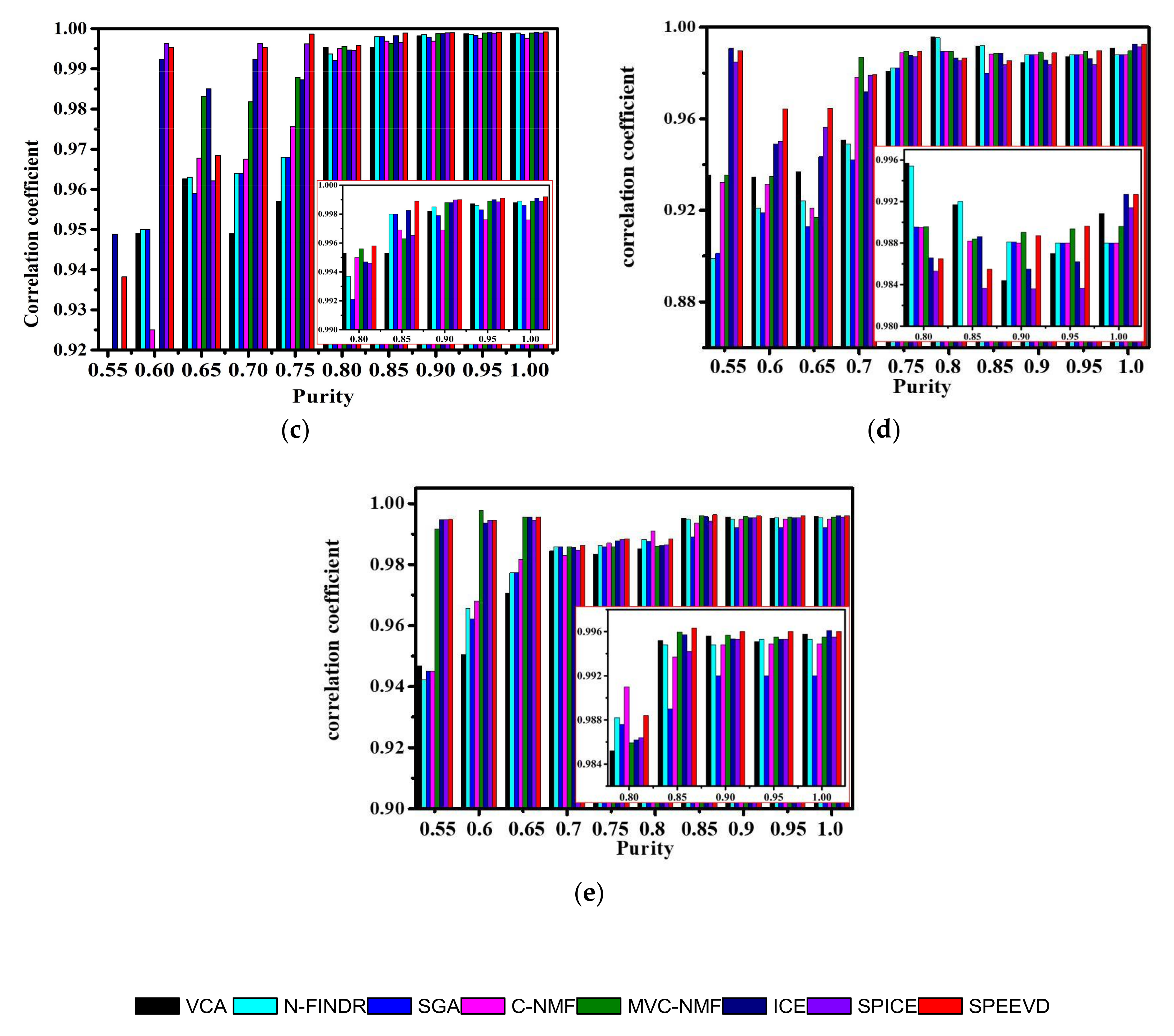


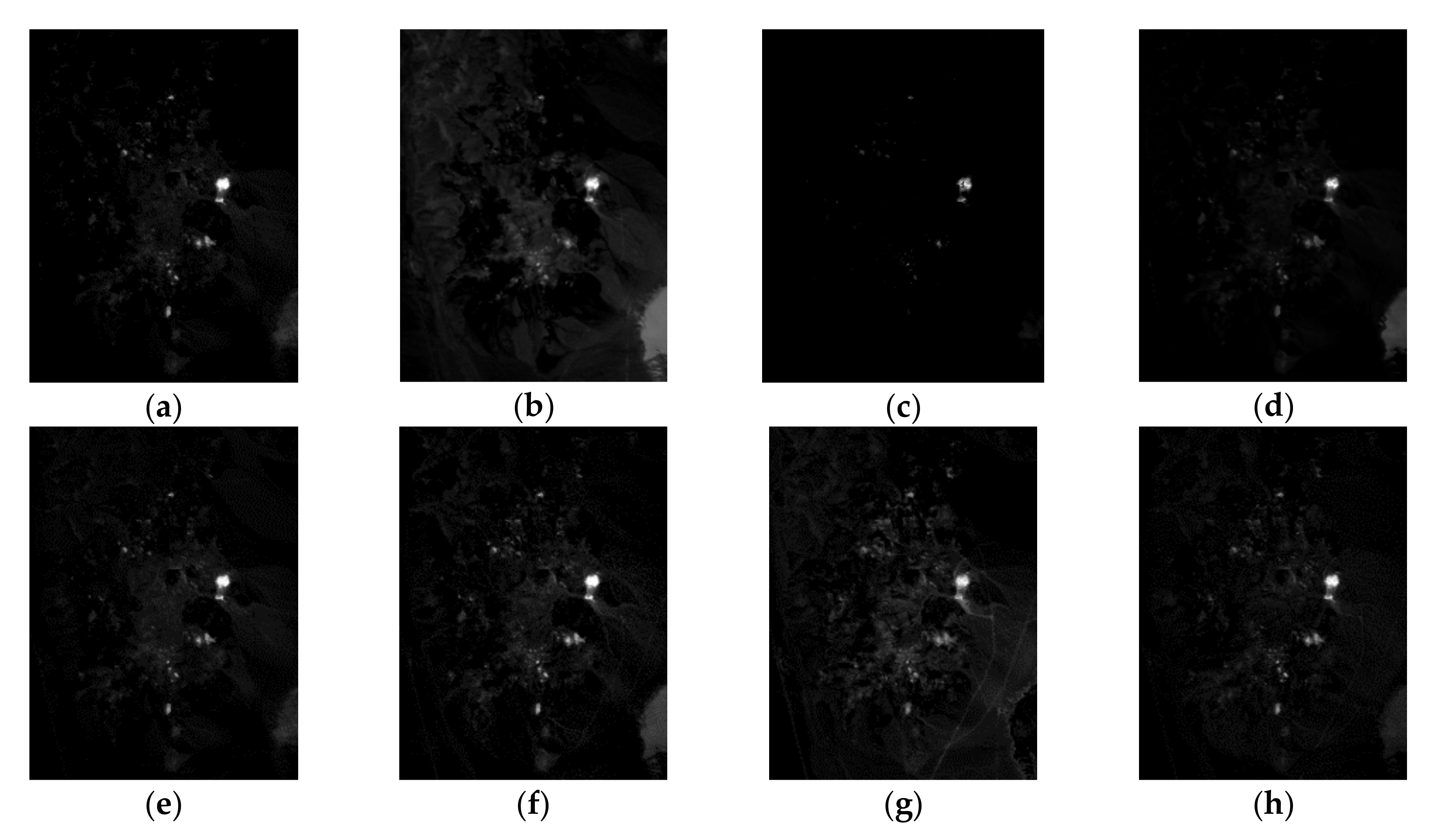

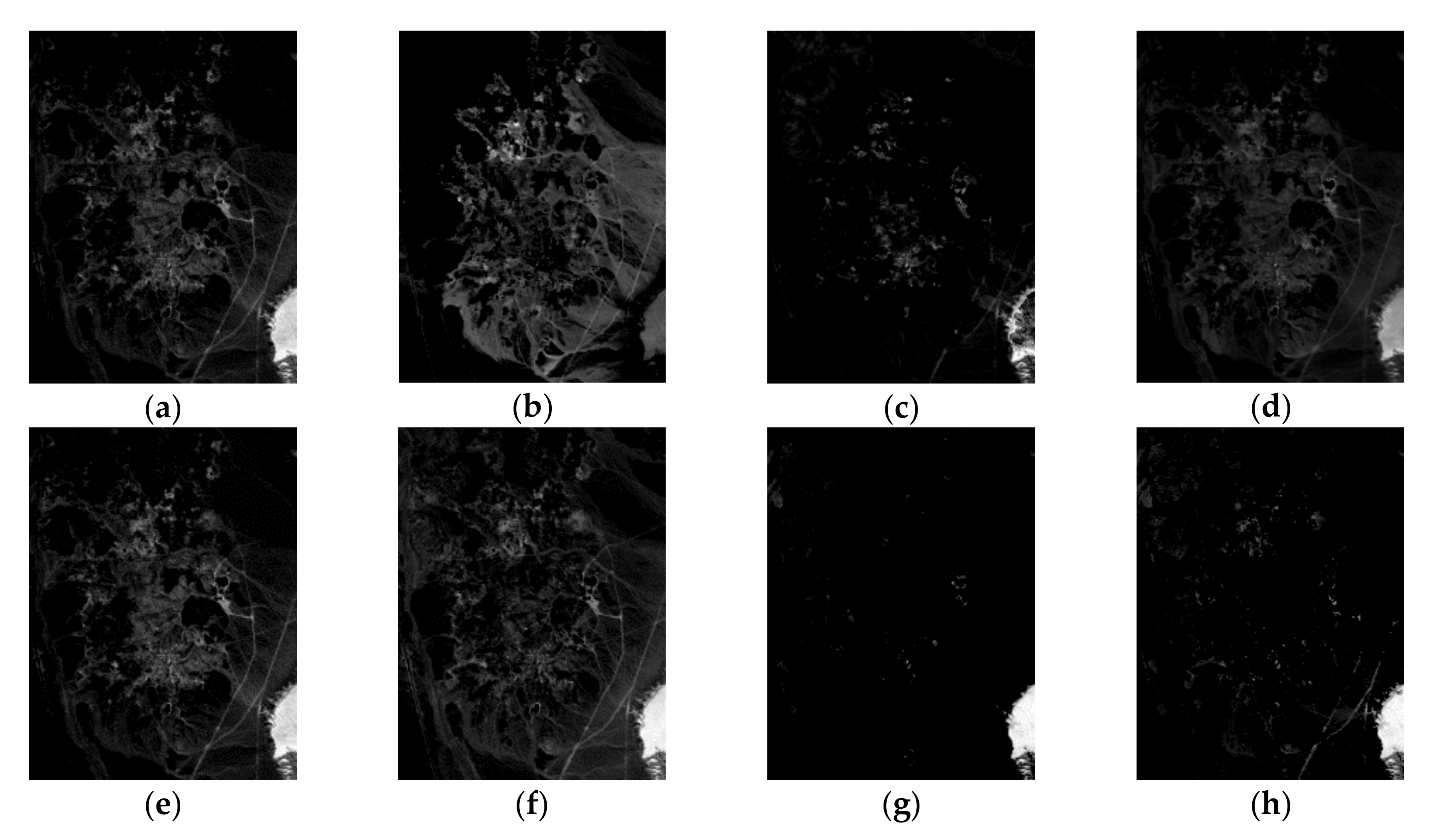
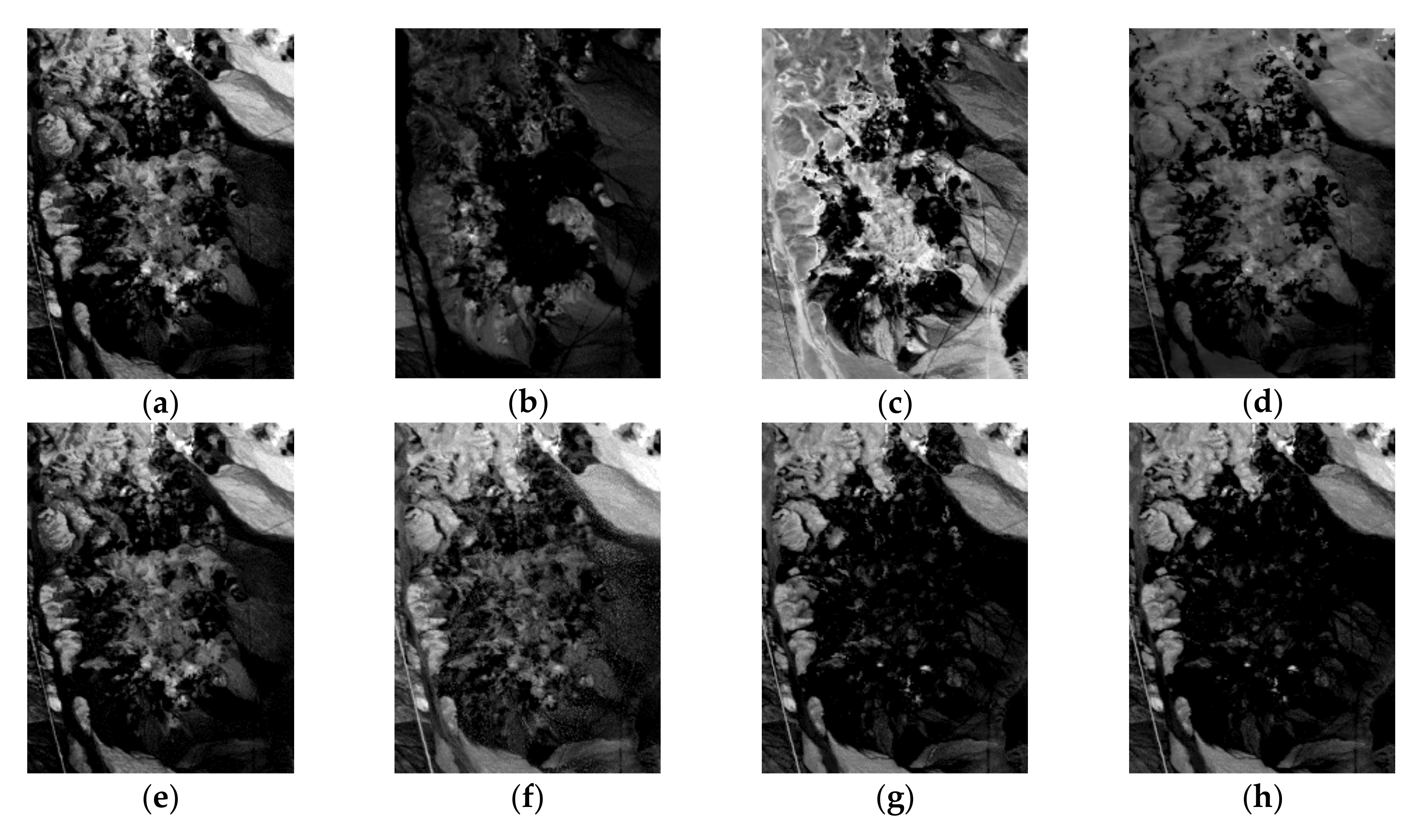
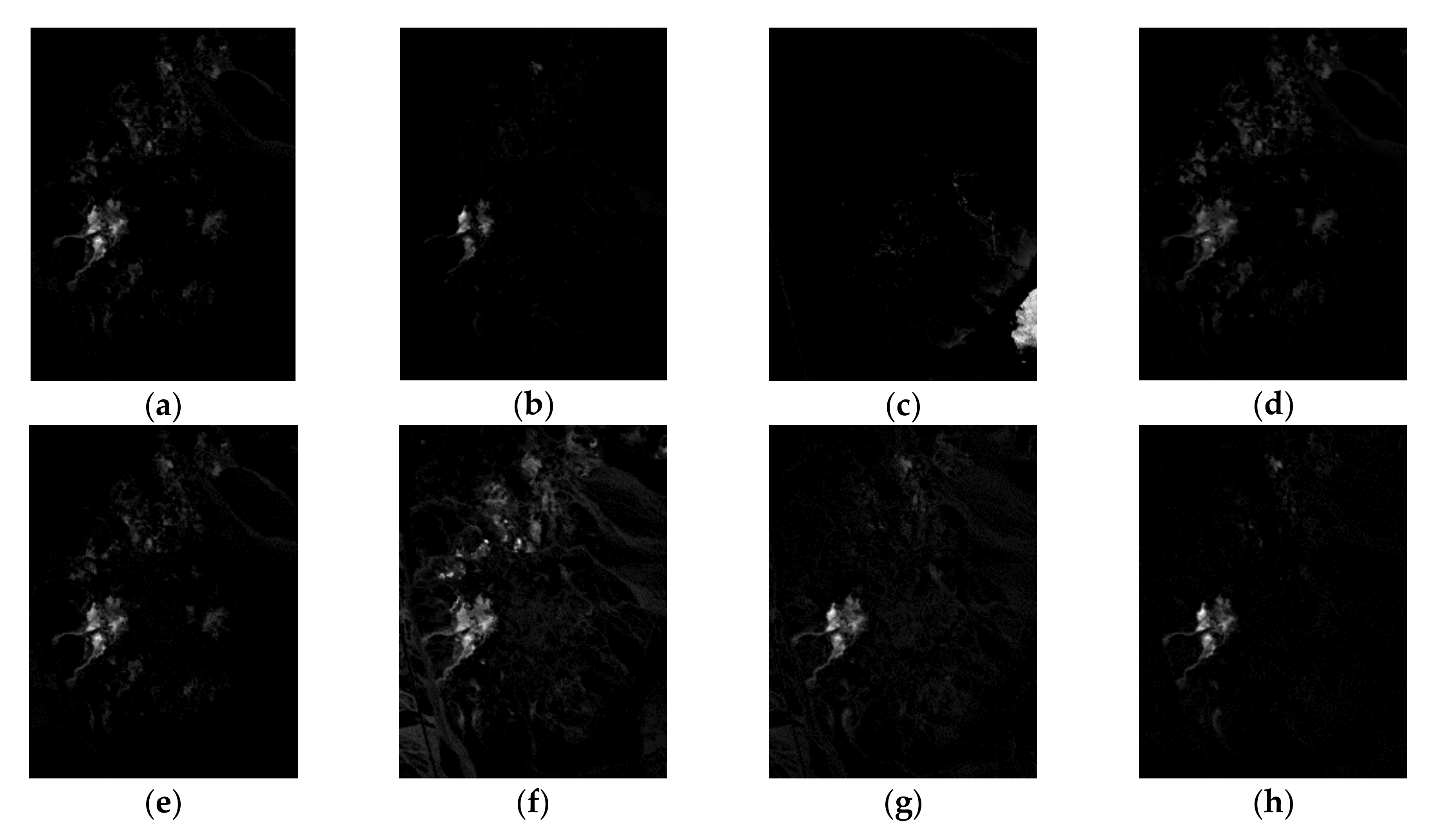
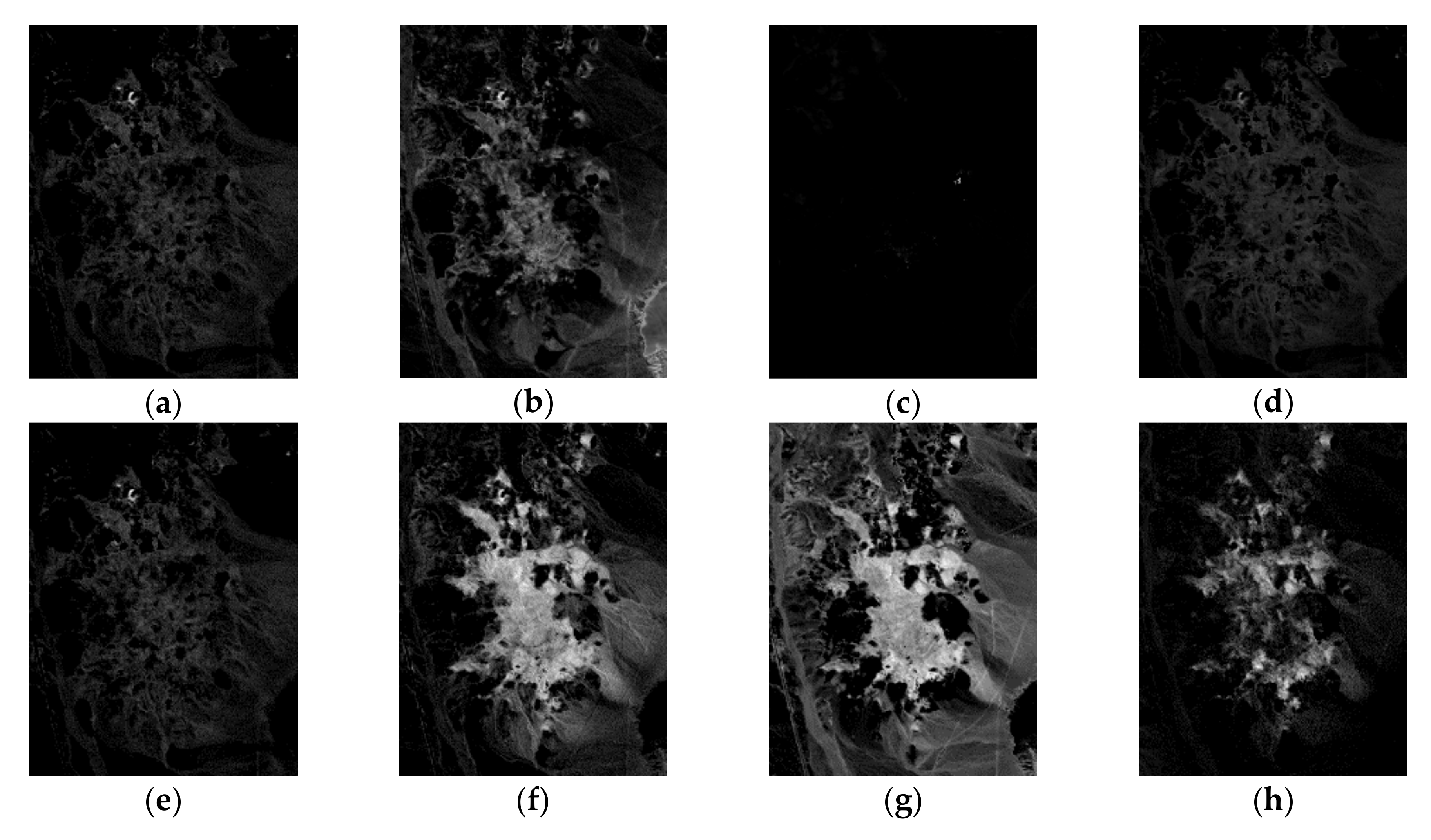

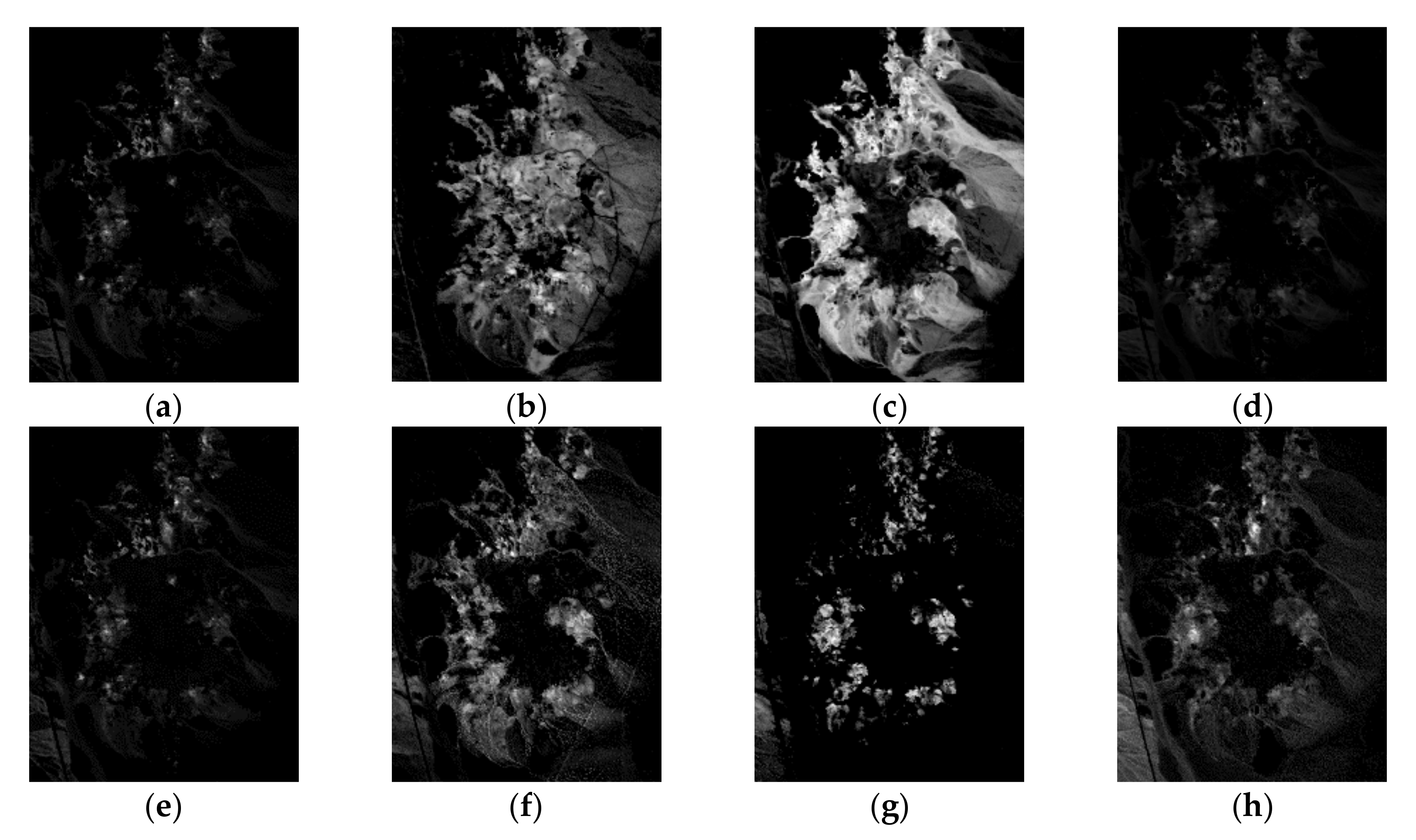

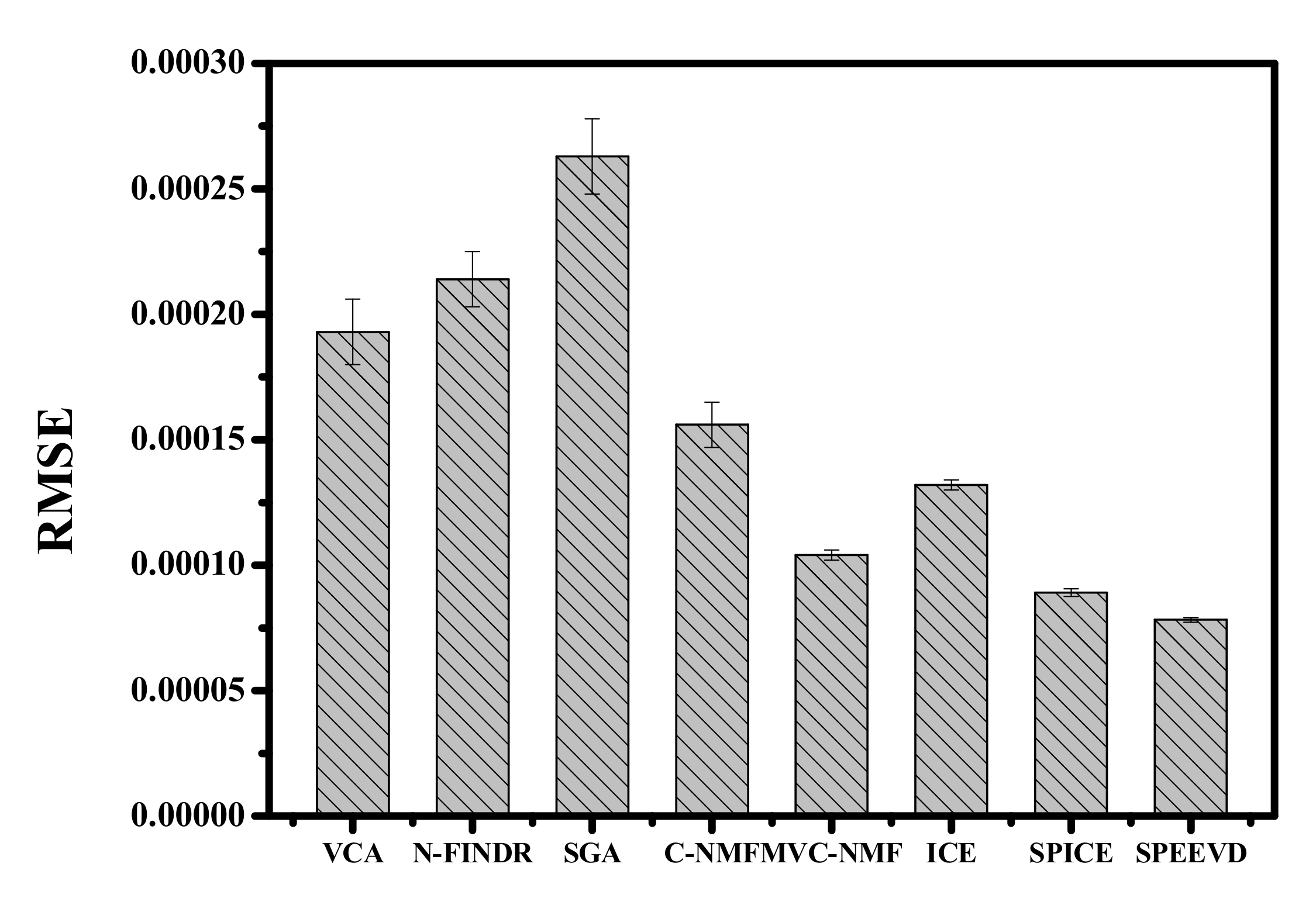



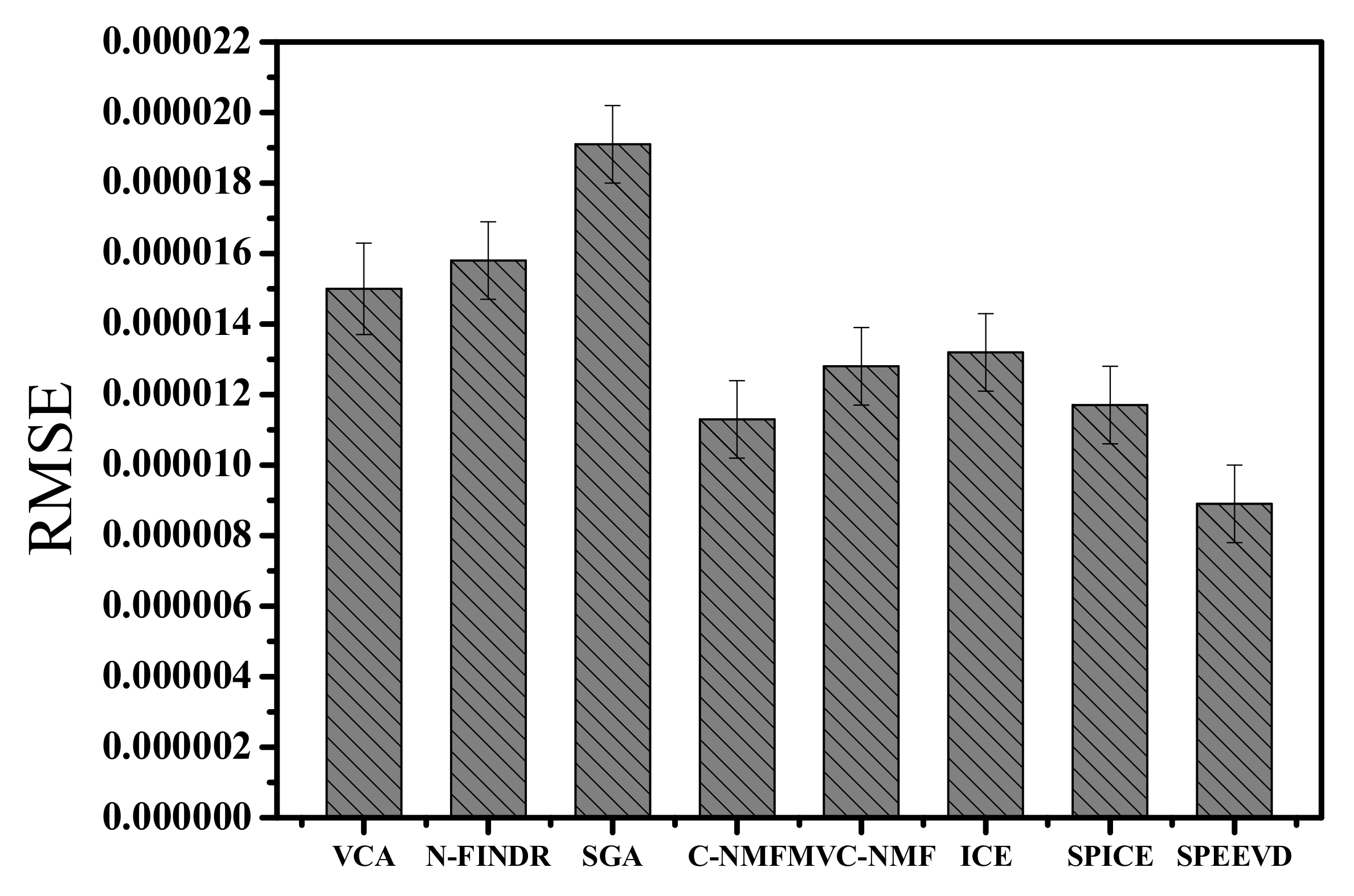
| Correlation | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| Coefficient | ||||||||
| Carnallite | 0.9987 | 0.9982 | 0.9972 | 0.9989 | 0.9989 | 0.9986 | 0.9988 | 0.9991 |
| Chlorite | 0.9979 | 0.9980 | 0.9980 | 0.9987 | 0.9983 | 0.9978 | 0.9992 | 0.9993 |
| Clinochlore | 0.9964 | 0.9889 | 0.9785 | 0.9970 | 0.9972 | 0.9970 | 0.9972 | 0.9974 |
| Clintonite | 0.9035 | 0.9045 | 0.9088 | 0.9840 | 0.9986 | 0.9269 | 0.9846 | 0.9981 |
| Corundum | 0.9971 | 0.9961 | 0.9689 | 0.9980 | 0.9978 | 0.9961 | 0.9970 | 0.9978 |
| Cos(SAD) | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| Carnallite | 0.9984 | 0.9983 | 0.9980 | 0.9991 | 0.9991 | 0.9984 | 0.9987 | 0.9993 |
| Chlorite | 0.9988 | 0.9981 | 0.9981 | 0.9993 | 0.9989 | 0.9984 | 0.9982 | 0.9994 |
| Clinochlore | 0.9998 | 0.9989 | 0.9894 | 0.9999 | 0.9999 | 0.9998 | 0.9998 | 0.9999 |
| Clintonite | 0.9962 | 0.9937 | 0.9656 | 0.9992 | 0.9999 | 0.9963 | 0.9965 | 0.9989 |
| Corundum | 0.9997 | 0.9989 | 0.9979 | 0.9998 | 0.9998 | 0.9996 | 0.9998 | 0.9999 |
| SID-SAD | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| Carnallite | 24.5356 | 24.5356 | 49.1908 | 22.0654 | 20.1549 | 46.6332 | 24.4198 | 22.8277 |
| Chlorite | 8.4406 | 8.6724 | 8.6744 | 5.6652 | 8.3581 | 8.3730 | 8.2069 | 5.4573 |
| Clinochlore | 0.1819 | 0.1994 | 1.3178 | 0.1540 | 0.1605 | 0.1701 | 0.1816 | 0.1514 |
| Clintonite | 26.0397 | 27.9036 | 19.5728 | 15.8640 | 14.7124 | 17.6472 | 16.2487 | 15.7286 |
| Corundum | 0.9892 | 2.3918 | 3.1188 | 0.5895 | 0.5878 | 0.5961 | 0.9701 | 0.5254 |
| Three Group of Parameters | |||
|---|---|---|---|
| μ | 0.00005 | 0.0001 | 0.0002 |
| T | 0.0001 | 0.0001 | 0.0001 |
| Estimated number | 5 | 5 | 5 |
| Method | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| Correlation | 0.8023 | 0.8157 | 0.7863 | 0.9682 | 0.9789 | 0.9436 | 0.9793 | 0.9813 |
| Coefficient |
| Method | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| Cos(SAD) | 0.9073 | 0.8961 | 0.8174 | 0.9523 | 0.9915 | 0.9668 | 0.9877 | 0.9937 |
| Method | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| SID-SAD | 12.9073 | 14.8157 | 20.7863 | 10.9682 | 10.9519 | 10.9668 | 10.9707 | 10.9376 |
| Time Cost | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| seconds | 3.08 | 112.3 | 3.45 | 31.4 | 45.36 | 19.3 | 39.5 | 47.74 |
| Time Cost | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| seconds | 18.46 | 712.1 | 25.2 | 313.6 | 458.6 | 334.2 | 398.3 | 506.8 |
| Time Cost | VCA | N-FINDR | SGA | C-NMF | MVC-NMF | ICE | SPICE | SPEEVD |
|---|---|---|---|---|---|---|---|---|
| seconds | 4.198 | 217.3 | 7.16 | 161.83 | 152.6 | 79.3 | 113.3 | 174.26 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Liu, J.; Yu, H. An Automatic Sparse Pruning Endmember Extraction Algorithm with a Combined Minimum Volume and Deviation Constraint. Remote Sens. 2018, 10, 509. https://doi.org/10.3390/rs10040509
Li H, Liu J, Yu H. An Automatic Sparse Pruning Endmember Extraction Algorithm with a Combined Minimum Volume and Deviation Constraint. Remote Sensing. 2018; 10(4):509. https://doi.org/10.3390/rs10040509
Chicago/Turabian StyleLi, Huali, Jun Liu, and Haicong Yu. 2018. "An Automatic Sparse Pruning Endmember Extraction Algorithm with a Combined Minimum Volume and Deviation Constraint" Remote Sensing 10, no. 4: 509. https://doi.org/10.3390/rs10040509