Biodegradation of Malachite Green in Milkfish Pond Sediments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
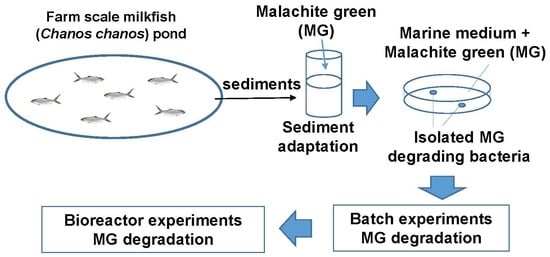
2.2. Sample Collection
2.3. Experimental Design
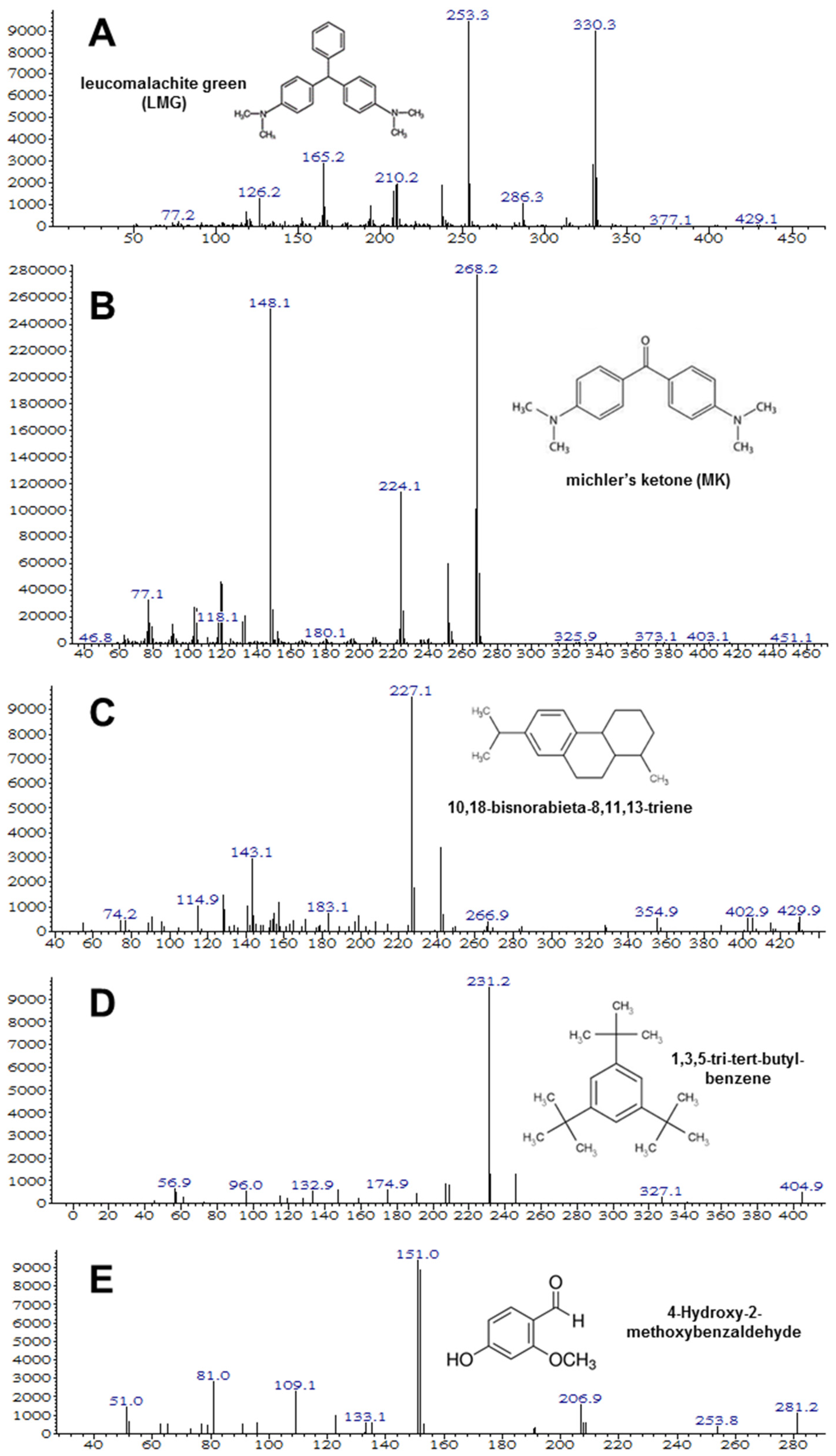
2.4. Analytical Methods
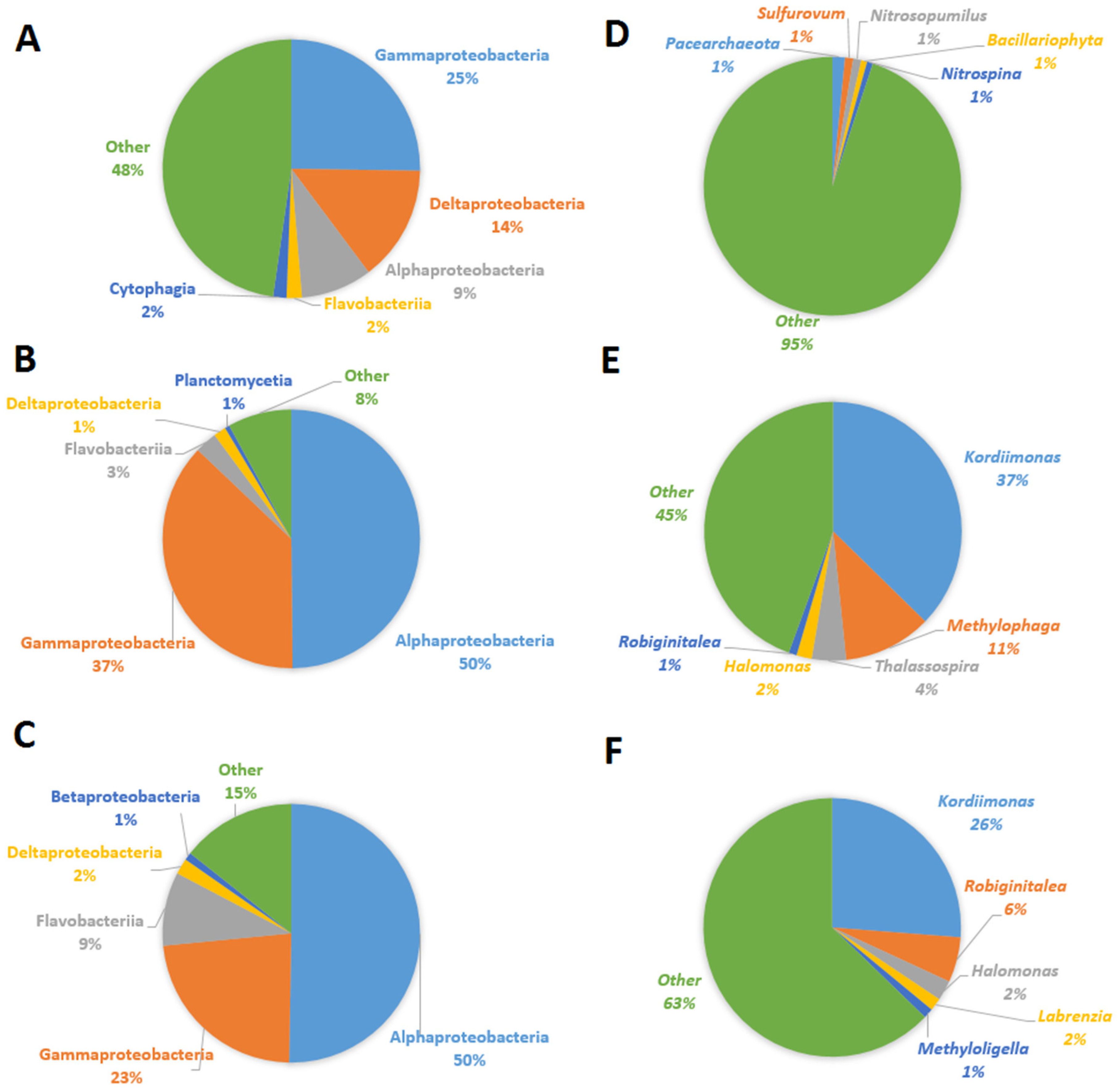
2.5. DNA Extraction, PCR, Illumina Sequencing, and Data Analysis
3. Results and Discussion
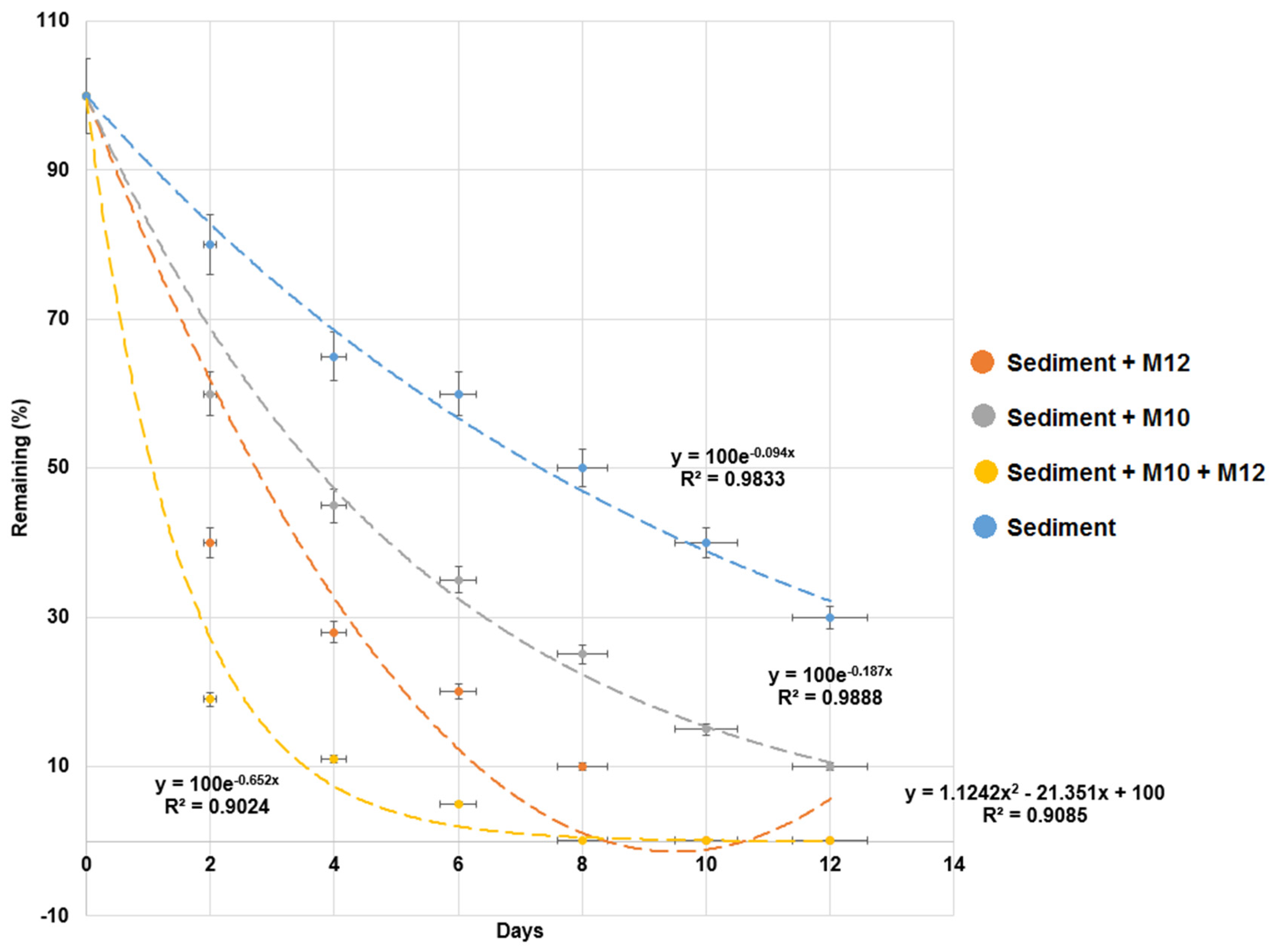
3.1. Effect of Different Conditions on MG Degradation in Milkfish Pond Sediments
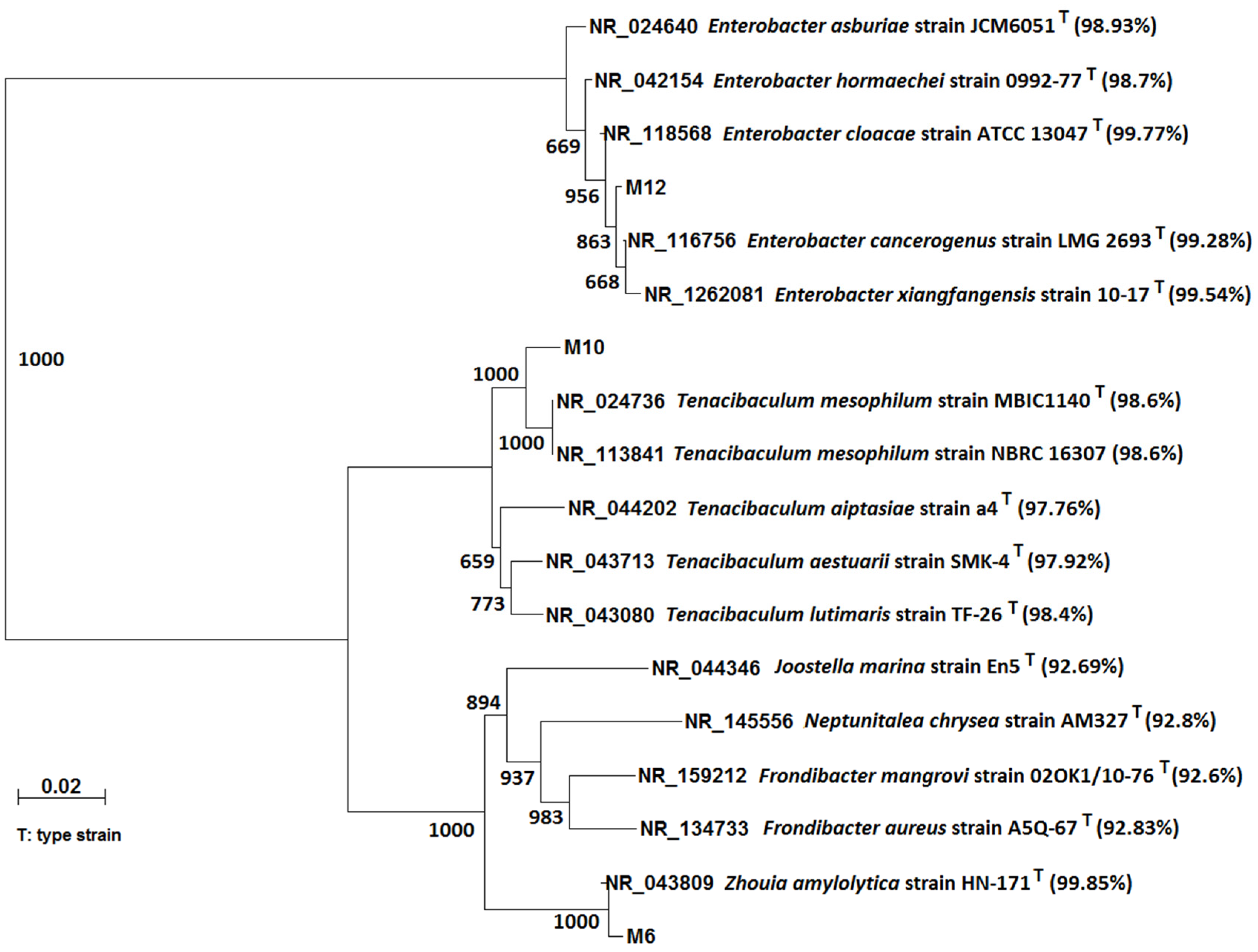
3.2. Isolation and Characterization of the MG-Degrading Bacteria
3.3. Test of MG Degradation Ability in Milkfish Pond Sediment
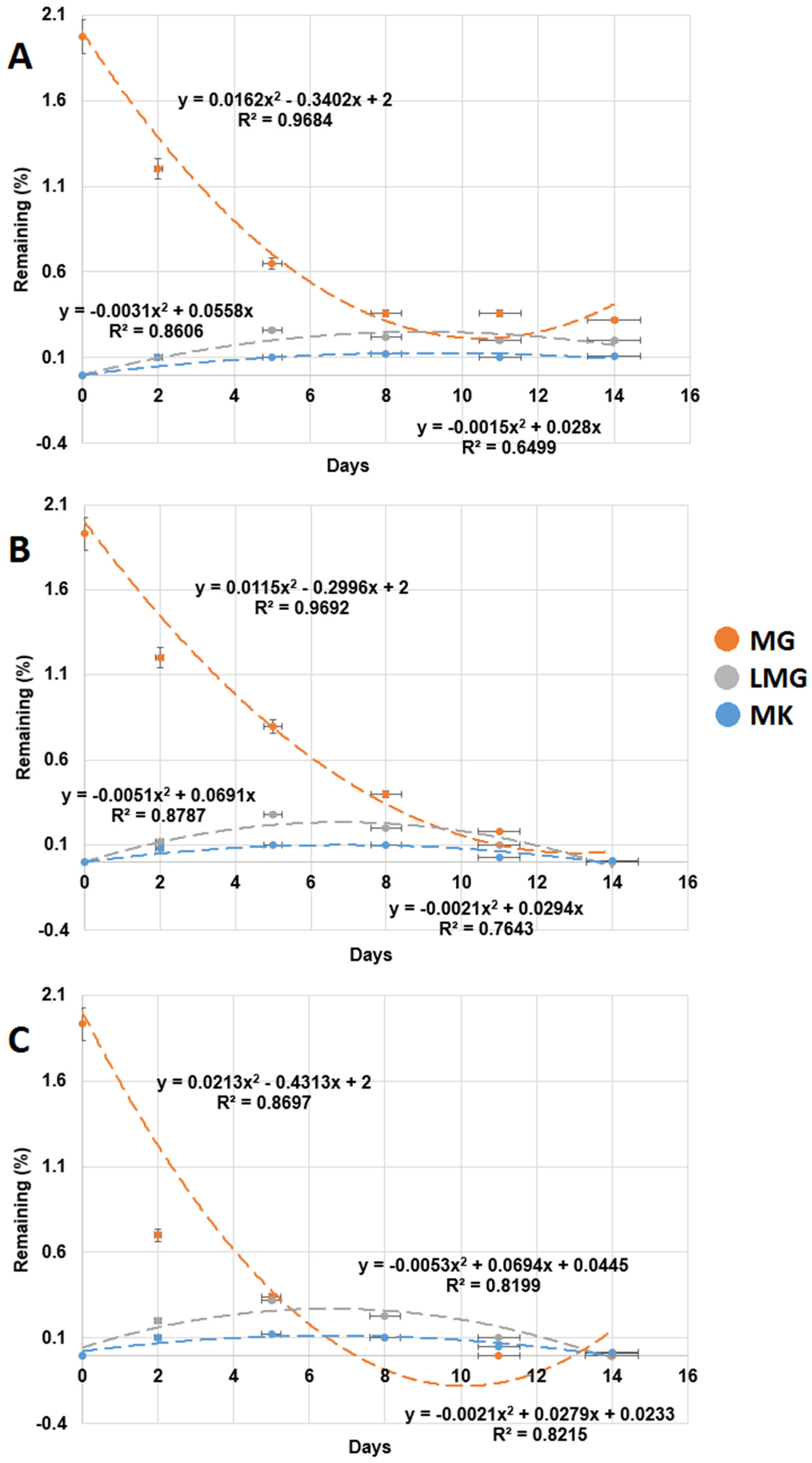
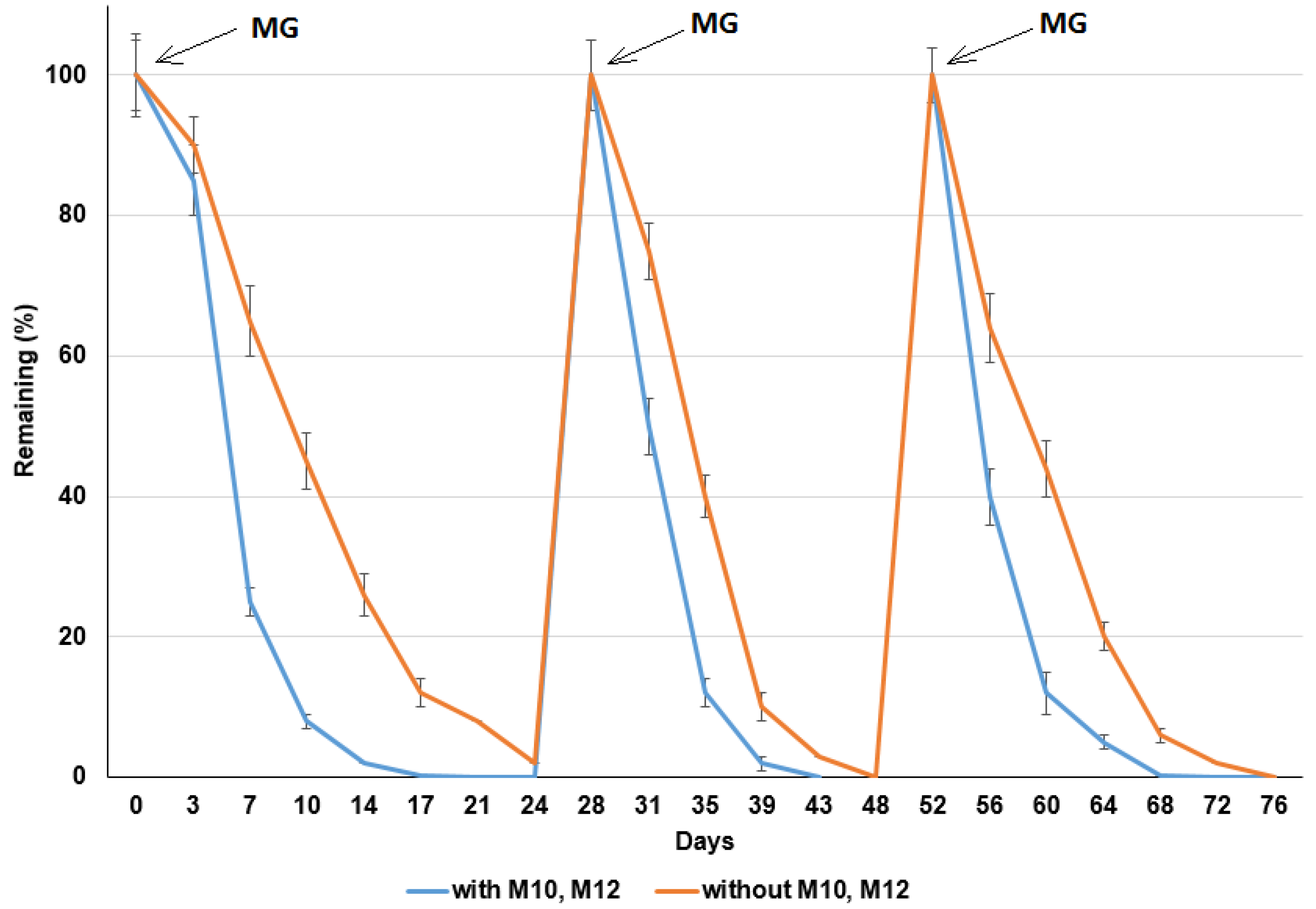
3.4. Repeated Addition of MG Influencing MG Degradation in the Sediments
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bagarinao, T. Systematics, distribution, genetics and life history of Milkfish Chanos Chanos. Environ. Biol. Fish. 1994, 39, 25–41. [Google Scholar] [CrossRef]
- Martinez, F.S.; Tseng, M.C.; Yeh, S.P. Milkfish (Chanos chanos) culture: Situations and trends. J. Fish. Soc. Taiwan. 2006, 33, 229–244. [Google Scholar]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef] [PubMed]
- Maalej-Kammoun, M.; Zouari-Mechichi, H.; Belbahri, L.; Woodward, S.; Mechichi, T. Malachite green decolourization and detoxification by the laccase from a newly isolated strain of Trametes sp. Int. Biodeterior. Biodegrad. 2009, 63, 600–606. [Google Scholar] [CrossRef]
- Srivastava, S.; Sinha, R.; Roy, D. Toxicological effects of malachite green. Aquat. Toxicol. 2004, 66, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Du, L.N.; Wang, S.; Li, G.; Wang, B.; Jia, X.M.; Zhao, Y.H.; Chen, Y.L. Biodegradation of malachite green by Pseudomonas sp. strain DY1 under aerobic condition: Characteristics, degradation products, enzyme analysis and phytotoxicity. Ecotoxicology 2011, 20, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, J.P.; Govindwar, S.P. Biotransformation of malachite green by Saccharomyces Cerevisiae MTCC 463. Yeast 2006, 23, 315–323. [Google Scholar] [CrossRef]
- Chang, C.J.; Doerge, D.R.; Cerniglia, C.E. Biotransformation of malachite green by the fungus Cunninghamella Elegans. Appl. Environ. Microbiol. 2001, 67, 4358–4360. [Google Scholar]
- Shanmugam, S.; Ulaganathan, P.; Swaminathan, K.; Sadhasivam, S.; Wu, Y.R. Enhanced biodegradation and detoxification of malachite green by Trichoderma asperellum laccase: Degradation pathway and product analysis. Int. Biodeterior. Biodegrad. 2018, 125, 258–268. [Google Scholar] [CrossRef]
- Banet, I.M.; Nigam, P.; Singh, D.; Marchant, R. Microbial decolorization of textile dye containing effluent a review. Bioresour. Technol. 1996, 58, 217–227. [Google Scholar] [CrossRef]
- Daneshvar, N.; Ayazloo, M.; Khataee, A.R.; Pourhassan, M. Biological decolourization of dye solution containing malachite green by microalgae Cosmarium sp. Bioresour. Technol. 2007, 98, 1176–1182. [Google Scholar] [CrossRef]
- Parshetti, G.; Kalme, S.; Saratale, G.; Govindwar, S. Biodegradation of malachite green by Kocuria rosea MTCC 15. Acta Chim. Slov. 2006, 53, 493–498. [Google Scholar]
- Jefferson, J.; Joseph, O.F. Decolorization of malachite green and crystal violet by waterborne pathogenic mycobacteria. Antimicrob. Agents Chemother. 2003, 47, 2323–2326. [Google Scholar]
- Chen, C.Y.; Kuo, J.T.; Cheng, C.Y.; Huang, Y.T.; Ho, I.H.; Chung, Y.C. Biological decolorization of dye solution containing malachite green by Pandoraea pulmonicola YC32 using a batch and continuous system. J. Hazard Mater. 2009, 172, 1439–1445. [Google Scholar] [CrossRef]
- Ayed, L.; Chaieb, K.; Cheref, A.; Bakhrouf, A. Biodegradation of triphenylmethane dye malachite green by Sphingomonas Paucimobilis. World J. Microbiol. Biotechnol. 2009, 25, 705–711. [Google Scholar] [CrossRef]
- Khataee, A.R.; Zarei, M.; Dehghan, G.; Ebadi, E.; Pourhassan, M. Biotreatment of a triphenylmethane dye solution using a Xanthophyta alga: Modeling of key factors by neural network. J. Taiwan Inst. Chem. Eng. 2011, 42, 380–386. [Google Scholar] [CrossRef]
- Tao, Y.; Wang, F.; Meng, L.; Guo, Y.; Han, M.; Li, J.; Sun, C.; Wang, S. Biological decolorization and degradation of malachite green by Pseudomonas sp. YB2: Process optimization and biodegradation pathway. Curr. Microbiol. 2017, 74, 1210–1215. [Google Scholar] [CrossRef]
- Jung, J.; Seo, H.; Lee, S.H.; Jeon, C.O.; Park, W. The effect of toxic malachite green on the bacterial community in Antarctic soil and the physiology of malachite green-degrading Pseudomonas sp. MGO. Appl. Microbiol. Biotechnol. 2013, 97, 4511–4521. [Google Scholar] [CrossRef]
- Qu, W.; Liu, T.; Wang, D.; Hong, G.; Zhao, J. Metagenomics-based discovery of malachite green-degradation gene families and enzymes from mangrove sediment. Front. Microbiol. 2018, 9, 2187. [Google Scholar] [CrossRef]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Tringe, S.G.; Alvarez-Cohen, L. High-Throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. MBio 2015, 6, e02288-14. [Google Scholar] [CrossRef]
- Huang, H.W.; Chang, B.V.; Cheng, C.H. Biodegradation of dibromodiphenyl ether in river sediment. Int. Biodeterior. Biodegrad. 2012, 68, 1–6. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Sáez-Plaza, P.; Navas, M.J.; Wybraniec, S.; Michałowski, T.; Asuero, A.G. An overview of the Kjeldahl method of nitrogen determination. Part II. Sample preparation, working scale, instrumental finish, and quality control. Crit. Rev. Anal. Chem. 2013, 43, 224–272. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association/American Water Works Association/Water Environment Federation: Washington, DC, USA, 2005. [Google Scholar]
- Gerbersdorf, S.U.; Jancke, T.; Westrich, B. Physico-chemical and biological sediment properties determining erosion resistance of contaminated riverine sediments–Temporal and vertical pattern at the Lauffen reservoir/River Neckar, Germany. Limnologica 2005, 35, 132–144. [Google Scholar] [CrossRef]
- Yang, C.W.; Tsai, L.L.; Chang, B.V. Anaerobic degradation of sulfamethoxazole in mangrove sediments. Sci. Total. Environ. 2018, 643, 1446–1455. [Google Scholar] [CrossRef]
- Wright, E.S. Using DECIPHER v2.0 to Analyze Big Biological Sequence Data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. v.1.0.12 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 4 January 2019).
- Ting, W.T.E.; Yuan, S.Y.; Wu, S.D.; Chang, B.V. Biodegradation of phenanthrene and pyrene by Ganoderma Lucidum. Int. Biodeterior. Biodegrad. 2011, 65, 238–242. [Google Scholar] [CrossRef]
- Farha, A.K.; Tr, T.; Purushothaman, A.; Salam, J.A.; Hatha, A.M. Phylogenetic diversity and biotechnological potentials of marine bacteria from continental slope of eastern Arabian Sea. J. Genet. Eng. Biotechnol. 2018, 16, 253–258. [Google Scholar] [CrossRef]
- Liu, Z.P.; Wang, B.J.; Dai, X.; Liu, X.Y.; Liu, S.J. Zhouia Amylolytica gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 2006, 56, 2825–2829. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Toranzo, A.E.; Magariños, B. Tenacibaculosis infection in marine fish caused by Tenacibaculum Maritimum: A review. Dis. Aquat. Org. 2006, 71, 255–266. [Google Scholar] [CrossRef]
- Ren, Y.; Zhou, Z.; Guo, X.; Li, Y.; Feng, L.; Wang, L. Complete genome sequence of Enterobacter Cloacae Subsp. Cloacae type strain ATCC 13047. J. Bacteriol. 2010, 192, 2463–2464. [Google Scholar]
- Pepper, L.L.; Gerba, C.P.; Gentry, T.J. Environmental microbiology. In Microorganisms and Organic Pollutants; Maier, M.M., Gentry, T.J., Eds.; Elsevier Inc.: Berkeley, CA, USA, 2015. [Google Scholar]
- Chang, B.V.; Liu, C.L.; Yuan, S.Y.; Cheng, C.Y.; Ding, W.H. Biodegradation of nonylphenol in mangrove sediment. Int. Biodeterior. Biodegrad. 2008, 61, 325–330. [Google Scholar] [CrossRef]
- Chaturvedi, V.; Verma, P. Biodegradation of malachite green by a novel copper-tolerant Ochrobactrum Pseudogrignonense strain GGUPV1 isolated from copper mine waste water. Bioresour. Bioprocess 2015, 2, 42–50. [Google Scholar] [CrossRef]
- Arun Prasad, A.S.; Satyanarayana, V.S.; Bhaskara Rao, K.V. Biotransformation of Direct Blue 1 by a moderately halophilic bacterium Marinobacter sp. strain HBRA and toxicity assessment of degraded metabolites. J. Hazard Mater. 2013, 262, 674–684. [Google Scholar] [CrossRef]
- Stolze, Y.; Eikmeyer, F.; Wibberg, D.; Brandis, G.; Karsten, C.; Krahn, I.; Schneiker-Bekel, S.; Viehöver, P.; Barsch, A.; Keck, M.; et al. IncP-1β plasmids of Comamonas sp. and Delftia sp. strains isolated from a wastewater treatment plant mediate resistance to and decolorization of the triphenylmethane dye crystal violet. Microbiology 2012, 158, 2060–2072. [Google Scholar] [CrossRef]
- Wang, J.; Qiao, M.; Wei, K.; Ding, J.; Liu, Z.; Zhang, K.Q.; Huang, X. Decolorizing activity of malachite green and its mechanisms involved in dye biodegradation by Achromobacter Xylosoxidans MG1. J. Mol. Microbiol. Biotechnol. 2011, 20, 220–227. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, J.H.; Zhou, Y.G.; Liu, H.C.; Liu, Z.P. Flavobacterium Caeni sp. nov., isolated from a sequencing batch reactor for the treatment of malachite green effluents. Int. J. Syst. Evol. Microbiol. 2010, 60, 417–421. [Google Scholar] [CrossRef]
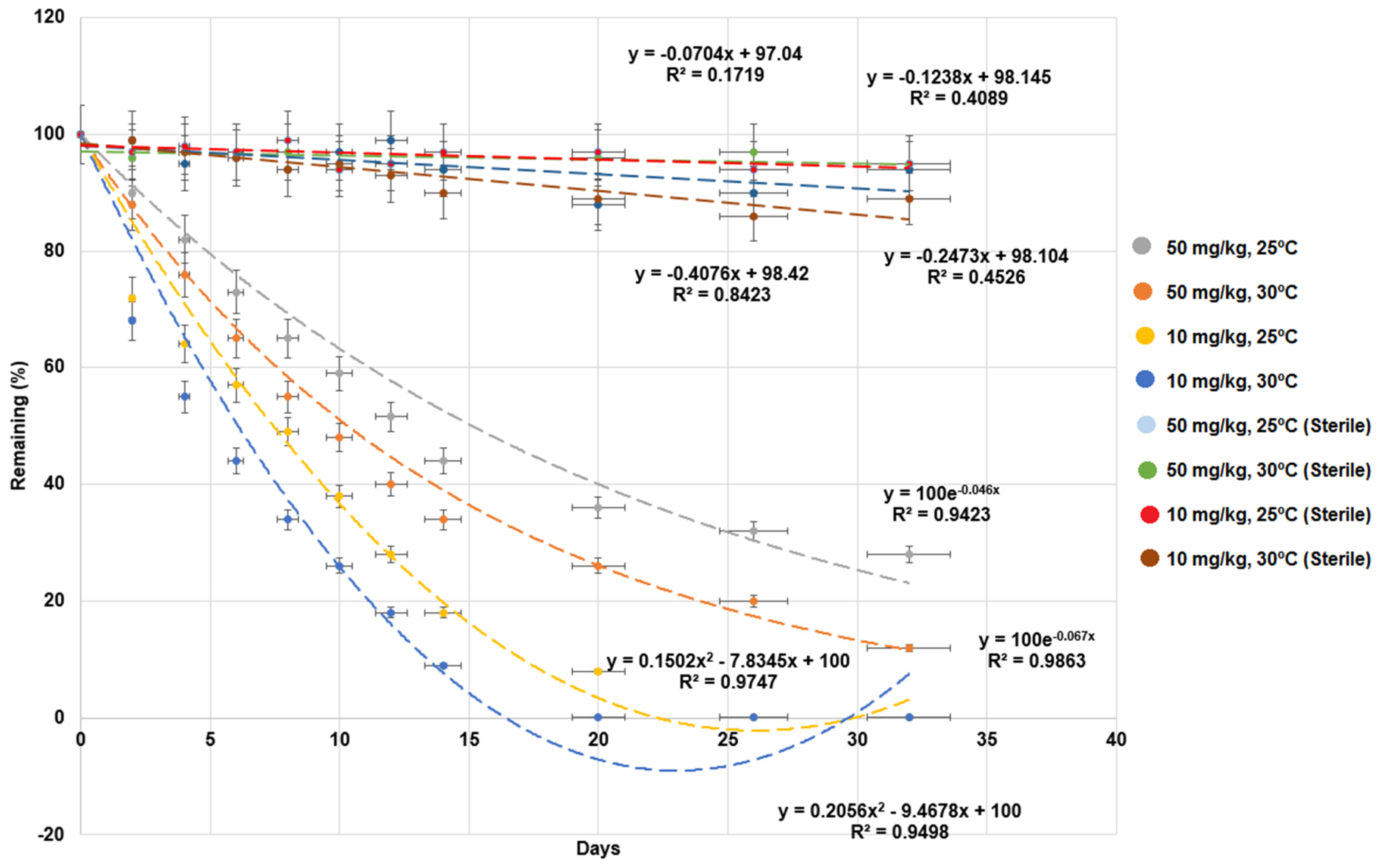


| Treatments | k (1/day) | t1/2 (day) | R2 |
|---|---|---|---|
| 10 mg/kg and 25 °C (Sterile) | 0.0046 ± 0.00023 | 107.40 ± 5.4 | 0.905 |
| 10 mg/kg and 30 °C (Sterile) | 0.0055 ± 0.00027 | 90.90 ± 4.5 | 0.940 |
| 50 mg/kg and 25 °C (Sterile) | 0.0029 ± 0.00014 | 172.80 ± 8.6 | 0.905 |
| 50 mg/kg and 30 °C (Sterile) | 0.0033 ± 0.00016 | 149.80 ± 7.5 | 0.955 |
| 10 mg/kg and 25 °C | 0.117 ± 0.006 | 6.0 ± 0.3 | 0.951 |
| 10 mg/kg and 30 °C | 0.151 ± 0.007 | 4.6 ± 0.2 | 0.962 |
| 50 mg/kg and 25 °C | 0.046 ± 0.002 | 15.1 ± 0.7 | 0.958 |
| 50 mg/kg and 30 °C | 0.067 ± 0.004 | 10.3 ± 0.5 | 0.987 |
| Strains | k (1/day) | t1/2 (day) | R2 |
|---|---|---|---|
| M6 | 0.169 ± 0.008 | 4.1 ± 0.205 | 0.958 |
| M10 | 0.273 ± 0.014 | 2.5 ± 0.123 | 0.991 |
| M12 | 0.377 ± 0.019 | 1.9 ± 0.095 | 0.951 |
| Treatment | With Sediment | Without Sediment | ||||
|---|---|---|---|---|---|---|
| k (1/day) | t1/2 (day) | R2 | k (1/day) | t1/2 (day) | R2 | |
| Without bacteria | 0.090 ± 0.005 | 7.7 ± 0.385 | 0.983 | __ | __ | __ |
| M10 | 0.182 ± 0.001 | 3.8 ± 0.193 | 0.959 | 0.273 ± 0.014 | 2.5 ± 0.123 | 0.991 |
| M12 | 0.292 ± 0.015 | 2.4 ± 0.127 | 0.966 | 0.377 ± 0.019 | 1.9 ± 0.095 | 0.951 |
| M10 and M12 | 0.538 ± 0.027 | 1.3 ± 0.065 | 0.958 | 0.693 ± 0.035 | 1.0 ± 0.042 | 0.964 |
| Treatments | With Degrading Bacteria | Without Degrading Bacteria | ||||
|---|---|---|---|---|---|---|
| k (1/day) | t1/2 (day) | R2 | k (1/day) | t1/2 (day) | R2 | |
| 1st addition | 0.289 ± 0.014 | 3.46 ± 0.173 | 0.961 | 0.128 ± 0.006 | 7.81 ± 0.391 | 0.974 |
| 2nd addition | 0.315 ± 0.016 | 3.17 ± 0.158 | 0.972 | 0.217 ± 0.011 | 4.6 ± 0.230 | 0.994 |
| 3rd addition | 0.347 ± 0.017 | 2.88 ± 0.144 | 0.964 | 0.257 ± 0.013 | 3.8 ± 0.194 | 0.953 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-W.; Chao, W.-L.; Hsieh, C.-Y.; Chang, B.-V. Biodegradation of Malachite Green in Milkfish Pond Sediments. Sustainability 2019, 11, 4179. https://doi.org/10.3390/su11154179
Yang C-W, Chao W-L, Hsieh C-Y, Chang B-V. Biodegradation of Malachite Green in Milkfish Pond Sediments. Sustainability. 2019; 11(15):4179. https://doi.org/10.3390/su11154179
Chicago/Turabian StyleYang, Chu-Wen, Wei-Liang Chao, Chi-Yen Hsieh, and Bea-Ven Chang. 2019. "Biodegradation of Malachite Green in Milkfish Pond Sediments" Sustainability 11, no. 15: 4179. https://doi.org/10.3390/su11154179