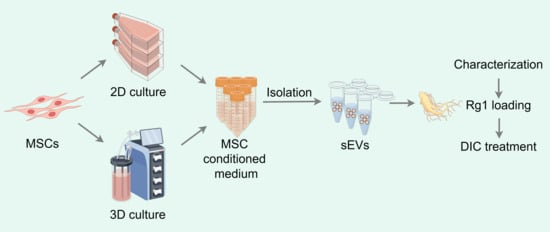
Mass Production of Rg1-Loaded Small Extracellular Vesicles Using a 3D Bioreactor System for Enhanced Cardioprotective Efficacy of Doxorubicin-Induced Cardiotoxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. SEM Imaging of Microcarriers
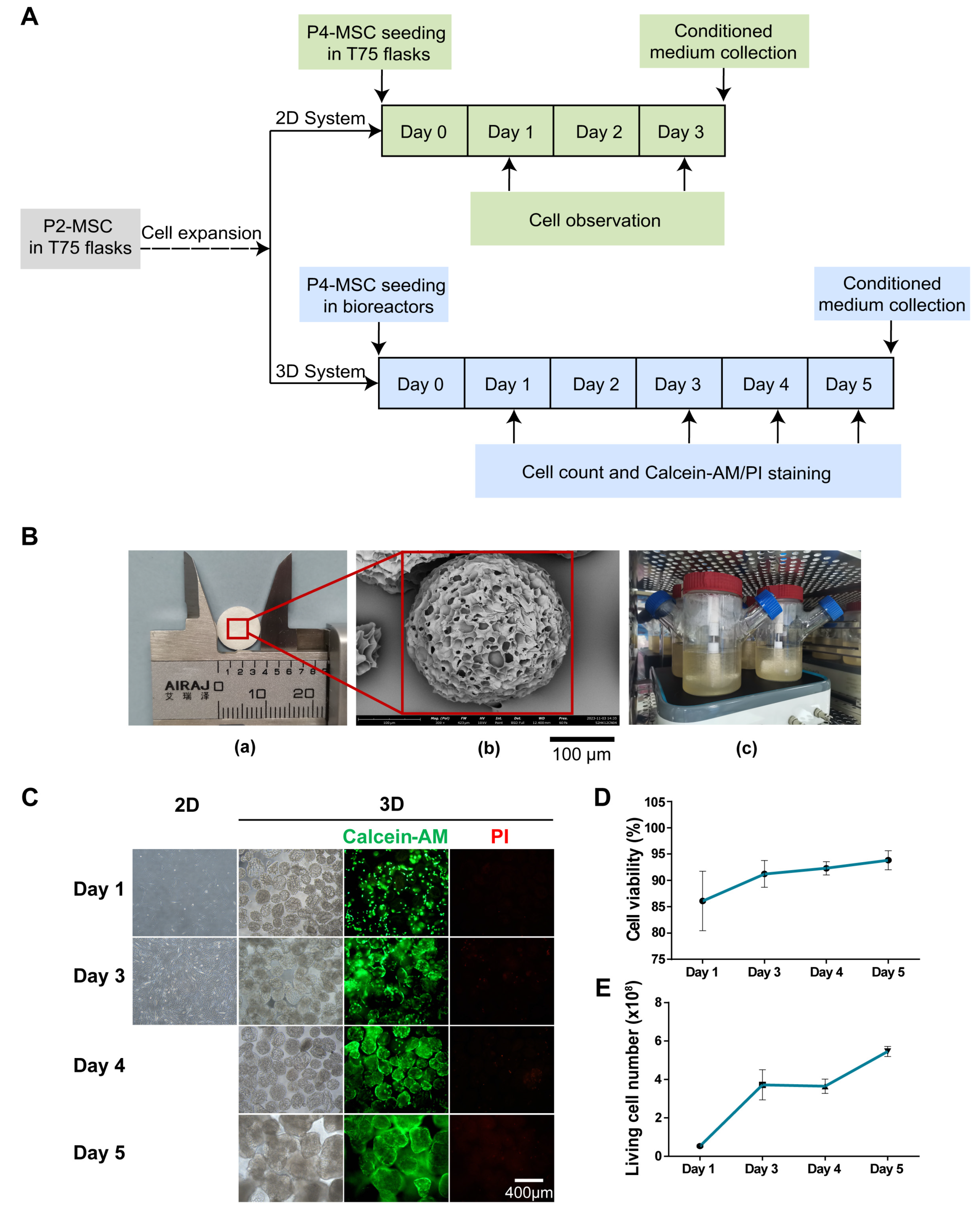
2.3. Scale-up Expansion of MSCs with Bioreactor System
2.4. Trypan Blue Staining
2.5. Isolation of sEVs
2.6. Characterization of sEVs
2.7. DOX Induced-Myocardial Injury Model
2.8. Drug Loading of sEVs
2.9. Drug Loading Efficiency of sEVs
2.10. Cell Viability Assay Using CCK-8
2.11. Flow Cytometry Assay
2.12. Live Cell Imaging and Analysis
2.13. sEV Uptake Essay
2.14. Statistical Analysis and Drawing
3. Results
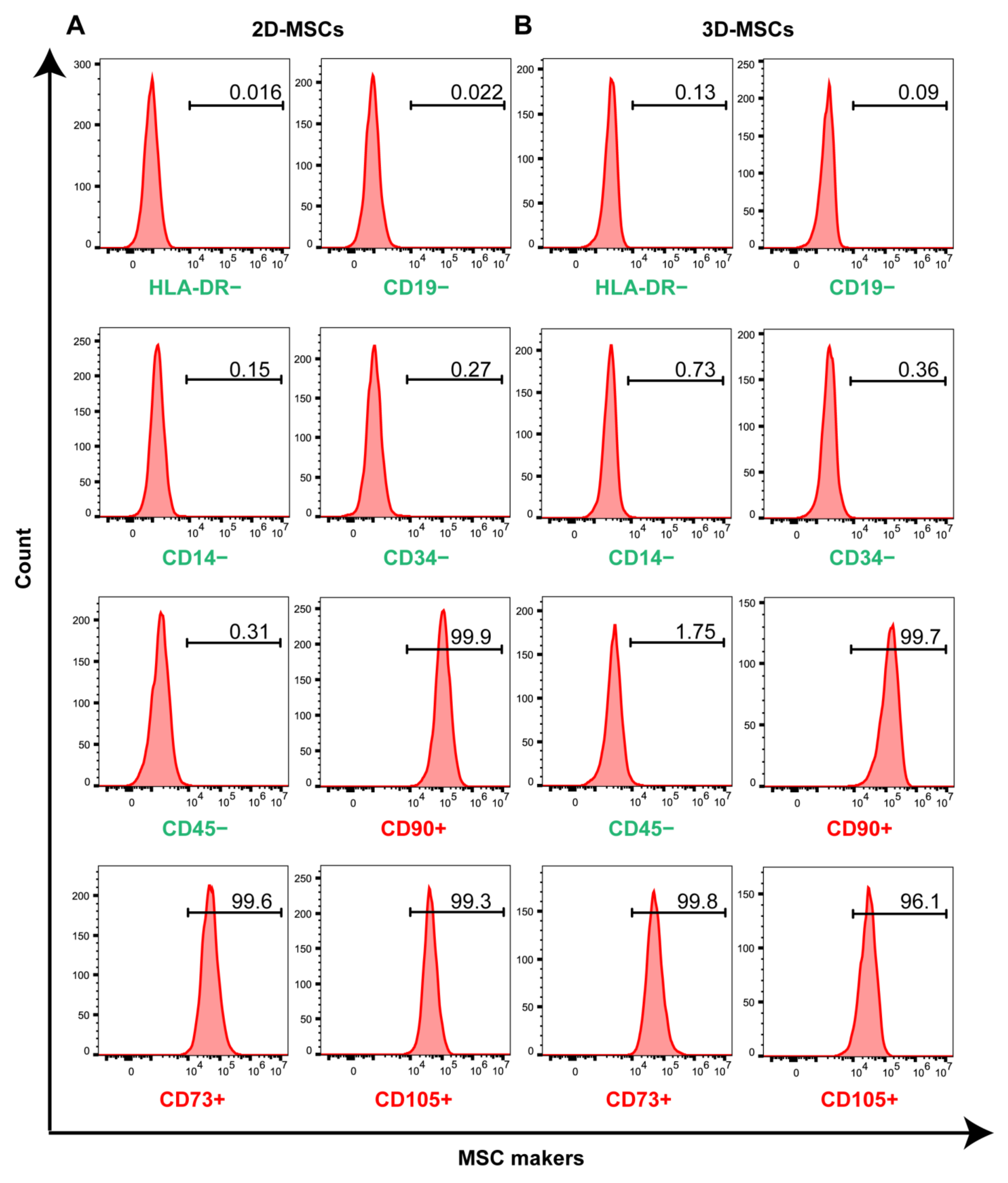
3.1. Three-Dimensional Rotating Bioreactor Scaled-up MSC Culture
3.2. Three-Dimensional Rotating Bioreactor Enabled Mass Production of sEVs
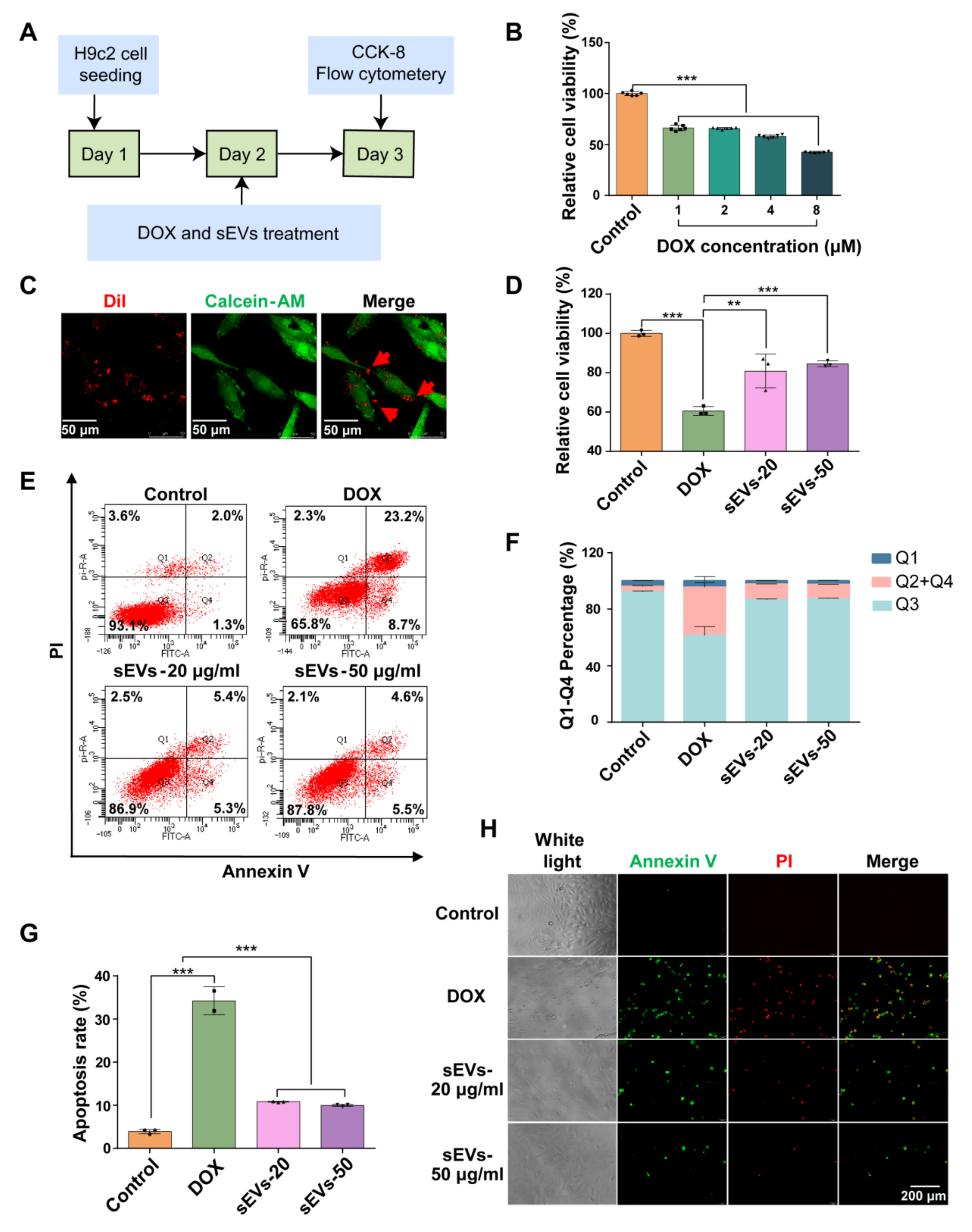
3.3. The 3D-sEVs Decreased H9c2 Cell Apoptosis and Protected H9c2 Cells from DOX-Induced Cardio Injury
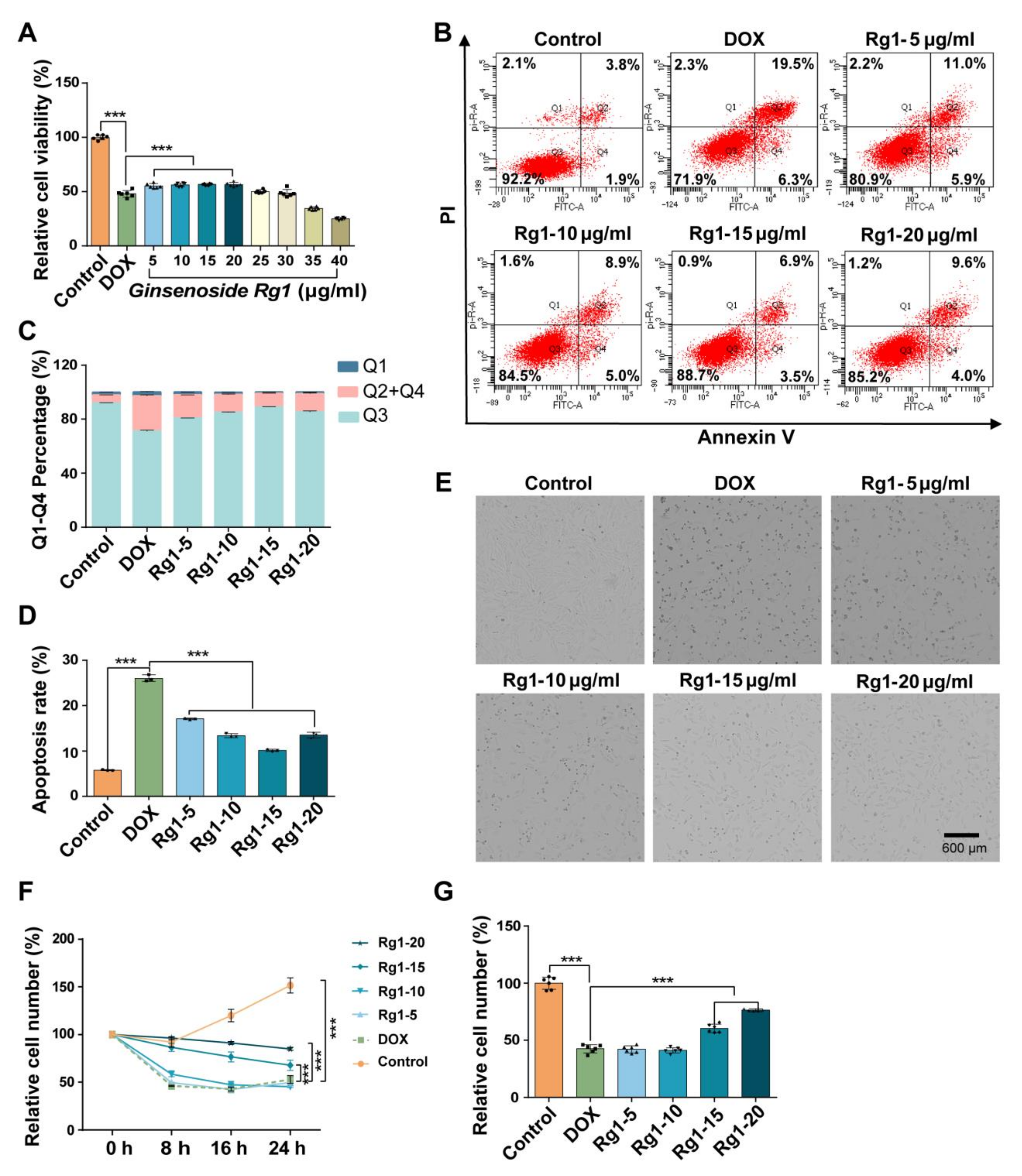
3.4. Ginsenoside Rg1 Decreased H9c2 Cell Apoptosis and Protected H9c2 Cells from DOX-Induced Cardio Injury
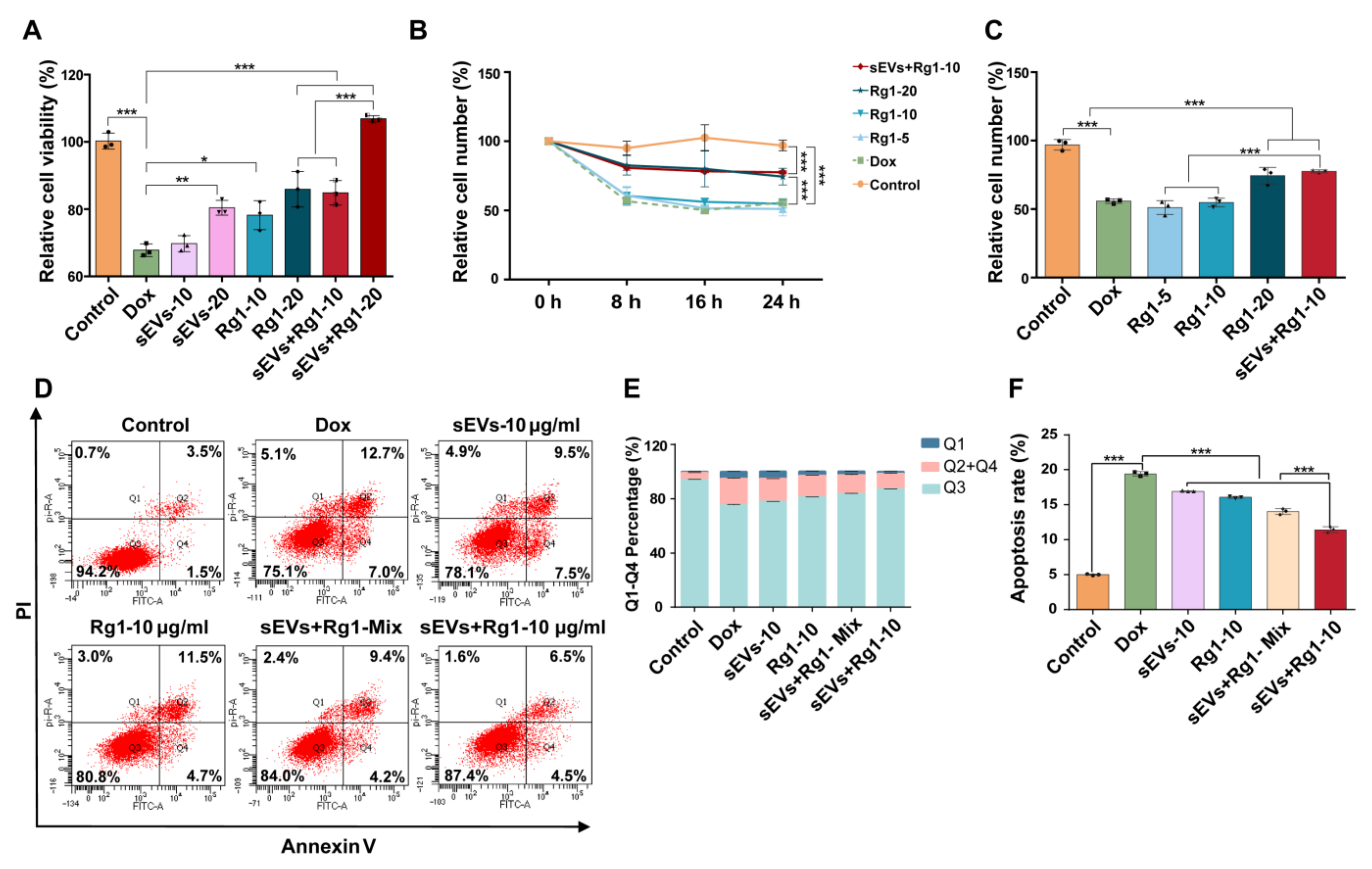
3.5. Rg1-Loaded sEVs Reduced DOX-Induced H9c2 Cell Damage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mattioli, R.; Ilari, A.; Colotti, B.; Mosca, L.; Fazi, F.; Colotti, G. Doxorubicin and other anthracyclines in cancers: Activity, chemoresistance and its overcoming. Mol. Aspects Med. 2023, 93, 101205. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, P.; Tabone, M.-D.; Mora, J.; Morland, B.; Jones, R.L. Risk-benefit of dexrazoxane for preventing anthracycline-related cardiotoxicity: Re-evaluating the European labeling. Future Oncol. 2018, 14, 2663–2676. [Google Scholar] [CrossRef] [PubMed]
- Christidi, E.; Brunham, L.R. Regulated cell death pathways in doxorubicin-induced cardiotoxicity. Cell Death Dis. 2021, 12, 339. [Google Scholar] [CrossRef]
- Zhang, H.; Wan, X.; Tian, J.; An, Z.; Liu, L.; Zhao, X.; Zhou, Y.; Zhang, L.; Ge, C.; Song, X. The therapeutic efficacy and clinical translation of mesenchymal stem cell-derived exosomes in cardiovascular diseases. Biomed. Pharmacother. 2023, 167, 115551. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Ghanam, J.; Chetty, V.K.; Zhu, X.; Liu, X.; Gelléri, M.; Barthel, L.; Reinhardt, D.; Cremer, C.; Thakur, B.K. Single Molecule Localization Microscopy for Studying Small Extracellular Vesicles. Small 2023, 19, 2205030. [Google Scholar] [CrossRef]
- Di, Y.; Wang, W.; Wang, Y.; Wang, J. Recent engineering advances of EVs for compounds, nucleic acids, and TCM delivery. Eur. J. Pharm. Sci. 2023, 190, 106584. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.-T.; Xiong, Y.-Y.; Ning, Y.; Tang, R.-J.; Xu, J.-Y.; Jiang, W.-Y.; Li, X.-S.; Zhang, L.-L.; Chen, C.; Pan, Q.; et al. Nicorandil-Pretreated Mesenchymal Stem Cell-Derived Exosomes Facilitate Cardiac Repair After Myocardial Infarction via Promoting Macrophage M2 Polarization by Targeting miR-125a-5p/TRAF6/IRF5 Signaling Pathway. Int. J. Nanomed. 2024, 19, 2005–2024. [Google Scholar] [CrossRef]
- Song, Y.; Wang, B.; Zhu, X.; Hu, J.; Sun, J.; Xuan, J.; Ge, Z. Human umbilical cord blood-derived MSCs exosome attenuate myocardial injury by inhibiting ferroptosis in acute myocardial infarction mice. Cell Biol. Toxicol. 2021, 37, 51–64. [Google Scholar] [CrossRef]
- Casajuana Ester, M.; Day, R.M. Production and Utility of Extracellular Vesicles with 3D Culture Methods. Pharmaceutics 2023, 15, 663. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Chen, J.-Y.; Tong, X.-M.; Mei, J.-G.; Chen, Y.-F.; Mou, X.-Z. Recent advances in the use of microcarriers for cell cultures and their ex vivo and in vivo applications. Biotechnol. Lett. 2019, 42, 1–10. [Google Scholar] [CrossRef]
- Xu, Z.-M.; Li, C.-B.; Liu, Q.-L.; Li, P.; Yang, H. Ginsenoside Rg1 Prevents Doxorubicin-Induced Cardiotoxicity through the Inhibition of Autophagy and Endoplasmic Reticulum Stress in Mice. Int. J. Mol. Sci. 2018, 19, 3658. [Google Scholar] [CrossRef]
- Fan, W.; Huang, Y.; Zheng, H.; Li, S.; Li, Z.; Yuan, L.; Cheng, X.; He, C.; Sun, J. Ginsenosides for the treatment of metabolic syndrome and cardiovascular diseases: Pharmacology and mechanisms. Biomed. Pharmacother. 2020, 132, 110915. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Lin, N.; Huang, H.; Zhang, X.; Cao, X.; Wang, Y.; Li, P. Ginsenoside Rg1 ameliorates cardiac oxidative stress and inflammation in streptozotocin-induced diabetic rats. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 1091–1103. [Google Scholar] [CrossRef]
- Baek, J.-S.; Yeon, W.-G.; Lee, C.-A.; Hwang, S.-J.; Park, J.-S.; Kim, D.-C.; Cho, C.-W. Preparation and characterization of mucoadhesive enteric-coating ginsenoside-loaded microparticles. Arch. Pharm. Res. 2014, 38, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mu, J.; Zheng, C.; Chen, X.; Guo, Z.; Huang, C.; Fu, Y.; Tian, G.; Shang, H.; Wang, Y. Systems-Pharmacology Dissection of Traditional Chinese Medicine Compound Saffron Formula Reveals Multi-scale Treatment Strategy for Cardiovascular Diseases. Sci. Rep. 2016, 6, 19809. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Gu, C.; Wang, W.; Wang, X.; Sun, J.; Zeng, B.; Chen, C.; Chang, W.; Ping, Y.; Ji, P.; et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol. Cancer 2020, 19, 117. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Kitakata, H.; Endo, J.; Ikura, H.; Moriyama, H.; Shirakawa, K.; Katsumata, Y.; Sano, M. Therapeutic Targets for DOX-Induced Cardiomyopathy: Role of Apoptosis vs. Ferroptosis. Int. J. Mol. Sci. 2022, 23, 1414. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, J.; Rahbarghazi, R.; Pezeshki, M.; Mazhar, M.; Yekani, F.; Khaksar, M.; Shokrollahi, E.; Amini, H.; Hashemzadeh, S.; Sokullu, S.E.; et al. Cardioprotective role of extracellular vesicles: A highlight on exosome beneficial effects in cardiovascular diseases. J. Cell. Physiol. 2019, 234, 21732–21745. [Google Scholar] [CrossRef]
- Aheget, H.; Tristán-Manzano, M.; Mazini, L.; Cortijo-Gutierrez, M.; Galindo-Moreno, P.; Herrera, C.; Martin, F.; Marchal, J.A.; Benabdellah, K. Exosome: A New Player in Translational Nanomedicine. J. Clin. Med. 2020, 9, 2380. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, L.; Fu, F.; Lai, Q.; Zhang, L.; Liu, T.; Yu, B.; Kou, J.; Li, F. Cardioprotective effect of ginsenoside Rb1 via regulating metabolomics profiling and AMP-activated protein kinase-dependent mitophagy. J. Ginseng Res. 2022, 46, 255–265. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Feng, R.; Sun, Y.; Chu, S.; Chen, J.; Ma, C.; Fu, J.; Zhao, Z.; Huang, M.; Shou, J.; et al. Simultaneous quantification of ginsenoside Rg1 and its metabolites by HPLC-MS/MS: Rg1 excretion in rat bile, urine and feces. Acta Pharm. Sin. B 2016, 6, 593–599. [Google Scholar] [CrossRef]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F.; Tsai, L.-C.; Hsieh, H.-Y.; Lu, K.-Y.; et al. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef]
- Bordin, A.; Chirivì, M.; Pagano, F.; Milan, M.; Iuliano, M.; Scaccia, E.; Fortunato, O.; Mangino, G.; Dhori, X.; De Marinis, E.; et al. Human platelet lysate-derived extracellular vesicles enhance angiogenesis through miR-126. Cell Prolif. 2022, 55, e13312. [Google Scholar] [CrossRef]
- Burnouf, T.; Chou, M.-L.; Lundy, D.J.; Chuang, E.-Y.; Tseng, C.-L.; Goubran, H. Expanding applications of allogeneic platelets, platelet lysates, and platelet extracellular vesicles in cell therapy, regenerative medicine, and targeted drug delivery. J. Biomed. Sci. 2023, 30, 79. [Google Scholar] [CrossRef]


| Terms | 3D Culture System | 2D Culture System # |
|---|---|---|
| Cell input | 8 × 107 cells | |
| Flasks | 4 spinner flasks | 132 T75 flasks |
| Conditioned medium | 2 × 103 mL | 1.58 × 103 mL |
| Total volume of sEVs | 3.3 × 103 μL | 3.96 × 103 μL |
| Average conc. of particles | 6.6 × 1011/mL | 3.35 × 1011/mL |
| Total particle number | 2.2 × 1012 | 1.33 × 1012 |
| Average conc. of protein | 0.53 mg/mL | 0.26 mg/mL |
| Total protein | 1.74 mg | 1.04 mg |
| Bench time * | 32 min | 264 min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, Y.; Zhao, S.; Fan, H.; Li, W.; Jiang, G.; Wang, Y.; Li, C.; Wang, W.; Wang, J. Mass Production of Rg1-Loaded Small Extracellular Vesicles Using a 3D Bioreactor System for Enhanced Cardioprotective Efficacy of Doxorubicin-Induced Cardiotoxicity. Pharmaceutics 2024, 16, 593. https://doi.org/10.3390/pharmaceutics16050593
Di Y, Zhao S, Fan H, Li W, Jiang G, Wang Y, Li C, Wang W, Wang J. Mass Production of Rg1-Loaded Small Extracellular Vesicles Using a 3D Bioreactor System for Enhanced Cardioprotective Efficacy of Doxorubicin-Induced Cardiotoxicity. Pharmaceutics. 2024; 16(5):593. https://doi.org/10.3390/pharmaceutics16050593
Chicago/Turabian StyleDi, Yunfeng, Shuang Zhao, Huilan Fan, Wei Li, Guangjian Jiang, Yong Wang, Chun Li, Wei Wang, and Jingyu Wang. 2024. "Mass Production of Rg1-Loaded Small Extracellular Vesicles Using a 3D Bioreactor System for Enhanced Cardioprotective Efficacy of Doxorubicin-Induced Cardiotoxicity" Pharmaceutics 16, no. 5: 593. https://doi.org/10.3390/pharmaceutics16050593