PEG–Lipid–PLGA Hybrid Particles for Targeted Delivery of Anti-Inflammatory Drugs
Abstract
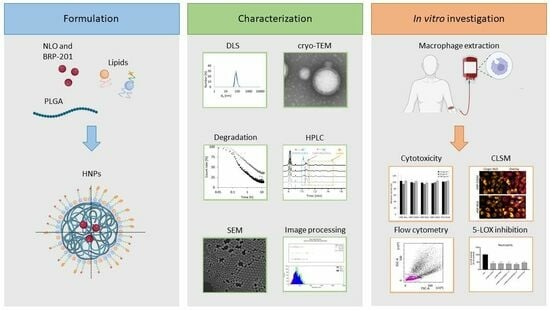
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Formulation of HNPs and NPs
2.2.2. Dynamic and Electrophoretic Light Scattering (DLS and ELS)
2.2.3. UV–Vis Spectroscopy
2.2.4. Scanning Electron Microscopy (SEM)
2.2.5. Analysis of the Particle Size Distributions in SEM Images
2.2.6. Cryo-Transmission Electron Microscopy (Cryo-TEM)
2.2.7. Size-Exclusion Chromatography (SEC)
2.2.8. High-Performance Liquid Chromatography (HPLC)
2.2.9. Isolation of Neutrophils and Macrophages
2.2.10. Differentiation and Polarization of Monocytes to Monocyte-Derived Macrophages
2.2.11. Incubation for Lipid Mediator (LM) Formation and LM Metabololipidomics via UPLC–MS-MS
2.2.12. Determination of Cell Viability
2.2.13. Evaluation of Cellular Uptake of NLO-Containing HNPs via Confocal Laser Scanning Microscopy (CLSM)
2.2.14. Evaluation of NLO Uptake via Flow Cytometry
2.2.15. Statistical Analysis
3. Results and Discussion
3.1. Formulation of Dye-Loaded PEG–Lipid–PLGA HNPs
3.2. Cytotoxicity and Uptake Studies of Dye-Loaded Particles in M0-MDMs
3.3. Dual-Loaded PEG–Lipid–PLGA HNPs with Different Sizes and Functionalities
3.3.1. Formulation of Dual-Loaded HNPs
3.3.2. HPLC Analysis of s-HNPs
3.3.3. Uptake Studies of Dual-Loaded Particles in M1-MDMs
3.3.4. Investigations Using FLAP-Dependent Cell-Based Bioactivity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, S.; Pandey, V.K.; Tewari, R.P.; Agarwal, V. Nanoparticle based drug delivery system: Advantages and applications. Indian J. Sci. Technol. 2011, 4, 177–180. [Google Scholar] [CrossRef]
- Wilczewska, A.Z.; Niemirowicz, K.; Markiewicz, K.H.; Car, H. Nanoparticles as drug delivery systems. Pharmacol. Rep. 2012, 64, 1020–1037. [Google Scholar] [CrossRef]
- Vauthier, C.; Ponchel, G. Polymer Nanoparticles for Nanomedicines; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Mamidi, N.; Velasco Delgadillo, R.M.; Barrera, E.V. Covalently functionalized carbon nano-onions integrated gelatin methacryloyl nanocomposite hydrogel containing γ-cyclodextrin as drug carrier for high-performance pH-triggered drug release. Pharmaceuticals 2021, 14, 291. [Google Scholar] [CrossRef]
- Hu, X.; Liu, S.; Zhou, G.; Huang, Y.; Xie, Z.; Jing, X. Electrospinning of polymeric nanofibers for drug delivery applications. J. Control. Release 2014, 185, 12–21. [Google Scholar] [CrossRef]
- DeLeon, V.H.; Nguyen, T.D.; Nar, M.; D’Souza, N.A.; Golden, T.D. Polymer nanocomposites for improved drug delivery efficiency. Mater. Chem. Phys. 2012, 132, 409–415. [Google Scholar] [CrossRef]
- Yingchoncharoen, P.; Kalinowski, D.S.; Richardson, D.R. Lipid-based drug delivery systems in cancer therapy: What is available and what is yet to come. Pharmacol. Rev. 2016, 68, 701–787. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Drug delivery systems: Entering the mainstream. Science 2004, 303, 1818–1822. [Google Scholar] [CrossRef]
- Mirakabad, F.S.T.; Nejati-Koshki, K.; Akbarzadeh, A.; Yamchi, M.R.; Milani, M.; Zarghami, N.; Zeighamian, V.; Rahimzadeh, A.; Alimohammadi, S.; Hanifehpour, Y.; et al. PLGA-based nanoparticles as cancer drug delivery systems. Asian Pac. J. Cancer 2014, 15, 517–535. [Google Scholar] [CrossRef] [PubMed]
- Elsabahy, M.; Wooley, K.L. Design of polymeric nanoparticles for biomedical delivery applications. Soc. Rev. 2012, 41, 2545–2561. [Google Scholar] [CrossRef] [PubMed]
- Hadinoto, K.; Sundaresan, A.; Cheow, W.S. Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013, 85, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Maurer, N.; Fenske, D.B.; Cullis, P.R. Developments in liposomal drug delivery systems. Expert Opin. Biol. Ther. 2001, 1, 923–947. [Google Scholar] [CrossRef]
- Shah, S.; Famta, P.; Raghuvanshi, R.S.; Singh, S.B.; Srivastava, S. Lipid polymer hybrid nanocarriers: Insights into synthesis aspects, characterization, release mechanisms, surface functionalization and potential implications. Colloids Interface Sci. Commun. 2022, 46, 100570. [Google Scholar] [CrossRef]
- Jain, S.; Kumar, M.; Kumar, P.; Verma, J.; Rosenholm, J.M.; Bansal, K.K.; Vaidya, A. Lipid–polymer hybrid nanosystems: A rational fusion for advanced therapeutic delivery. J. Funct. Biomater. 2023, 14, 437. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, G.; Krishna, K.V.; Dubey, S.K.; Taliyan, R. PEGylated Polymer–Lipid Hybrid Nanoparticles to Enhance In Vivo Exposure and Uptake of Repaglinide in Brain Cells to Treat Diabetes-Linked Neurodegenerative Disorders. ACS Appl. Nano Mater. 2023, 6, 3497–3512. [Google Scholar] [CrossRef]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly (ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. 2010, 49, 6288–6308. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.X.; Karnik, R.; Wang, A.Z.; Alexis, F.; Levy-Nissenbaum, E.; Hong, S.; Langer, R.S.; Farokhzad, O.C. Targeted nanoparticles for cancer therapy. Nano Today 2007, 2, 14–21. [Google Scholar] [CrossRef]
- Rasiel, A.; Sheskin, T.; Bergelson, L.; Domb, A.J. Phospholipid coated poly (lactic acid) microspheres for the delivery of LHRH analogues. Polym. Adv. Technol. 2002, 13, 127–136. [Google Scholar] [CrossRef]
- Zhang, L.; Chan, J.M.; Gu, F.X.; Rhee, J.-W.; Wang, A.Z.; Radovic-Moreno, A.F.; Alexis, F.; Langer, R.; Farokhzad, O.C. Self-assembled lipid—Polymer hybrid nanoparticles: A robust drug delivery platform. ACS Nano 2008, 2, 1696–1702. [Google Scholar] [CrossRef]
- Shi, J.-J.; Xiao, Z.-Y.; Votruba, A.R.; Vilos, C.; Farokhzad, O.C. Differentially charged hollow core/shell lipid-polymer-lipid hybrid nanoparticle for small interfering RNA delivery. Angew. Chem. Int. Ed. 2011, 50, 7027. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, M.; Gong, P.; Jia, D.; Zhang, P.; Shi, B.; Sheng, Z.; Ma, Y.; Cai, L. Indocyanine green-loaded biodegradable tumor targeting nanoprobes for in vitro and in vivo imaging. Biomaterials 2012, 33, 5603–5609. [Google Scholar] [CrossRef]
- Zaveri, T.D.; Lewis, J.S.; Dolgova, N.V.; Clare-Salzler, M.J.; Keselowsky, B.G. Integrin-directed modulation of macrophage responses to biomaterials. Biomaterials 2014, 35, 3504–3515. [Google Scholar] [CrossRef]
- Manzari, M.T.; Shamay, Y.; Kiguchi, H.; Rosen, N.; Scaltriti, M.; Heller, D.A. Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater. 2021, 6, 351–370. [Google Scholar] [CrossRef]
- Fujiwara, N.; Kobayashi, K. Macrophages in inflammation. Inflamm. Allergy Drug Targets 2005, 4, 281–286. [Google Scholar] [CrossRef]
- Evans, J.F.; Ferguson, A.D.; Mosley, R.T.; Hutchinson, J.H. What’s all the FLAP about?: 5-lipoxygenase-activating protein inhibitors for inflammatory diseases. Trends Pharmacol. Sci. 2008, 29, 72–78. [Google Scholar] [CrossRef]
- Gür, Z.T.; Çalışkan, B.; Garscha, U.; Olgaç, A.; Schubert, U.S.; Gerstmeier, J.; Werz, O.; Banoglu, E. Identification of multi-target inhibitors of leukotriene and prostaglandin E2 biosynthesis by structural tuning of the FLAP inhibitor BRP-7. Eur. J. Med. Chem. 2018, 150, 876–899. [Google Scholar] [CrossRef]
- Kretzer, C.; Jordan, P.M.; Bilancia, R.; Rossi, A.; Maz, T.G.; Banoglu, E.; Schubert, U.S.; Werz, O. Shifting the biosynthesis of leukotrienes toward specialized pro-resolving mediators by the 5-lipoxygenase-activating protein (FLAP) antagonist BRP-201. J. Inflamm. Res. 2022, 15, 911–925. [Google Scholar] [CrossRef]
- Koeberle, A.; Werz, O. Natural products as inhibitors of prostaglandin E2 and pro-inflammatory 5-lipoxygenase-derived lipid mediator biosynthesis. Biotechnol. Adv. 2018, 36, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- Kretzer, C.; Shkodra, B.; Klemm, P.; Jordan, P.M.; Schröder, D.; Cinar, G.; Vollrath, A.; Schubert, S.; Nischang, I.; Hoeppener, S. Ethoxy acetalated dextran-based nanocarriers accomplish efficient inhibition of leukotriene formation by a novel FLAP antagonist in human leukocytes and blood. Cell. Mol. Life Sci. 2022, 79, 40. [Google Scholar] [CrossRef] [PubMed]
- Banoglu, E.; Çelikoğlu, E.; Völker, S.; Olgaç, A.; Gerstmeier, J.; Garscha, U.; Çalışkan, B.; Schubert, U.S.; Carotti, A.; Macchiarulo, A.; et al. 4,5-Diarylisoxazol-3-carboxylic acids: A new class of leukotriene biosynthesis inhibitors potentially targeting 5-lipoxygenase-activating protein (FLAP). Eur. J. Med. Chem. 2016, 113, 1–10. [Google Scholar] [CrossRef]
- Chan, J.M.; Zhang, L.; Yuet, K.P.; Liao, G.; Rhee, J.-W.; Langer, R.; Farokhzad, O.C. PLGA–lecithin–PEG core–shell nanoparticles for controlled drug delivery. Biomaterials 2009, 30, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Bradski, G. The openCV library. Dr. Dobb’s J. Softw. Tools 2000, 25, 120–123. [Google Scholar]
- Pace, S.; Pergola, C.; Dehm, F.; Rossi, A.; Gerstmeier, J.; Troisi, F.; Pein, H.; Schaible, A.M.; Weinigel, C.; Rummler, S. Androgen-mediated sex bias impairs efficiency of leukotriene biosynthesis inhibitors in males. J. Clin. Investig. 2017, 127, 3167–3176. [Google Scholar] [CrossRef]
- Werz, O.; Gerstmeier, J.; Libreros, S.; De la Rosa, X.; Werner, M.; Norris, P.C.; Chiang, N.; Serhan, C.N. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat. Commun. 2018, 9, 59. [Google Scholar] [CrossRef]
- Werz, O.; Bürkert, E.; Samuelsson, B.; Rådmark, O.; Steinhilber, D. Activation of 5-lipoxygenase by cell stress is calcium independent in human polymorphonuclear leukocytes. Blood 2002, 99, 1044–1052. [Google Scholar] [CrossRef]
- Mohanty, A.; Uthaman, S.; Park, I.-K. Utilization of polymer-lipid hybrid nanoparticles for targeted anti-cancer therapy. Molecules 2020, 25, 4377. [Google Scholar] [CrossRef]
- Muljajew, I.; Huschke, S.; Ramoji, A.; Cseresnyés, Z.; Hoeppener, S.; Nischang, I.; Foo, W.; Popp, J.; Figge, M.T.; Weber, C.; et al. Stealth effect of short polyoxazolines in graft copolymers: Minor changes of backbone end group determine liver cell-type specificity. ACS Nano 2021, 15, 12298–12313. [Google Scholar] [CrossRef]
- Yu, S.S.; Lau, C.M.; Thomas, S.N.; Jerome, W.G.; Maron, D.J.; Dickerson, J.H.; Hubbell, J.A.; Giorgio, T.D. Size-and charge-dependent non-specific uptake of PEGylated nanoparticles by macrophages. Int. J. Nanomed. 2012, 7, 799–813. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Larsson, M.; Hill, A.; Duffy, J. Suspension stability; why particle size, zeta potential and rheology are important. Annu. Trans. Nord. Rheol. Soc. 2012, 20, 209–214. [Google Scholar]
- Behnke, M.; Klemm, P.; Dahlke, P.; Shkodra, B.; Beringer-Siemers, B.; Czaplewska, J.A.; Stumpf, S.; Jordan, P.M.; Schubert, S.; Hoeppener, S.; et al. Ethoxy acetalated dextran nanoparticles for drug delivery: A comparative study of formulation methods. Int. J. Pharm. X 2023, 5, 100173. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.L.; Román, J.S. Biodegradable Systems in Tissue Engineering and Regenerative Medicine; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Fredenberg, S.; Wahlgren, M.; Reslow, M.; Axelsson, A. The mechanisms of drug release in poly (lactic-co-glycolic acid)-based drug delivery systems—A review. Int. J. Pharm. 2011, 415, 34–52. [Google Scholar] [CrossRef]
- Zhang, K.; Tang, X.; Zhang, J.; Lu, W.; Lin, X.; Zhang, Y.; Tian, B.; Yang, H.; He, H. PEG–PLGA copolymers: Their structure and structure-influenced drug delivery applications. J. Control. Release 2014, 183, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Perinelli, R.; Cespi, M.; Bonacucina, G.; Palmieri, G.F. PEGylated polylactide (PLA) and poly (lactic-co-glycolic acid)(PLGA) copolymers for the design of drug delivery systems. J. Pharm. Investig. 2019, 49, 443–458. [Google Scholar] [CrossRef]
- Iversen, T.-G.; Skotland, T.; Sandvig, K. Endocytosis and intracellular transport of nanoparticles: Present knowledge and need for future studies. Nano Today 2011, 6, 176–185. [Google Scholar] [CrossRef]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, A.; Schallon, A.; Pietsch, C.; Schubert, S.; Nomoto, T.; Matsumoto, Y.; Kataoka, K.; Schubert, U.S. A toolbox of differently sized and labeled PMMA nanoparticles for cellular uptake investigations. Soft Matter 2013, 9, 99–108. [Google Scholar] [CrossRef]
- Sahin, A.; Esendagli, G.; Yerlikaya, F.; Caban-Toktas, S.; Yoyen-Ermis, D.; Horzum, U.; Aktas, Y.; Khan, M.; Couvreur, P.; Capan, Y. A small variation in average particle size of PLGA nanoparticles prepared by nanoprecipitation leads to considerable change in nanoparticles’ characteristics and efficacy of intracellular delivery. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Dhakar, R.C.; Maurya, S.D.; Sagar, B.P.; Bhagat, S.; Prajapati, S.K.; Jain, C.P. Variables influencing the drug entrapment efficiency of microspheres: A pharmaceutical review. Pharm. Lett. 2010, 2, 102–116. [Google Scholar]
- Mohammad, K.; Reineke, J.J. Quantitative detection of PLGA nanoparticle degradation in tissues following intravenous administration. Mol. Pharm. 2013, 10, 2183–2189. [Google Scholar] [CrossRef]
- Dunne, M.; Corrigan, O.; Ramtoola, Z. Influence of particle size and dissolution conditions on the degradation properties of polylactide-co-glycolide particles. Biomaterials 2000, 21, 1659–1668. [Google Scholar] [CrossRef]
- Tsarenko, E.; Schubert, U.S.; Nischang, I. Nanoparticle Formulation Composition Analysis by Liquid Chromatography on Reversed-Phase Monolithic Silica. Anal. Chem. 2023, 95, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Mousli, Y.; Brachet, M.; Chain, J.L.; Ferey, L. A rapid and quantitative reversed-phase HPLC-DAD/ELSD method for lipids involved in nanoparticle formulations. J. Pharm. Biomed. Anal. 2022, 220, 115011. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, C.; Lu, T.; Deiss, A.; Vuolo, K.; Klein, L.; Rustandi, R.R.; Loughney, J.W. Determination of lipid content and stability in lipid nanoparticles using ultra high-performance liquid chromatography in combination with a Corona Charged Aerosol Detector. Electrophoresis 2022, 43, 1091–1100. [Google Scholar] [CrossRef]
- Chono, S.; Tanino, T.; Seki, T.; Morimoto, K. Uptake characteristics of liposomes by rat alveolar macrophages: Influence of particle size and surface mannose modification. J. Pharm. Pharmacol. 2007, 59, 75–80. [Google Scholar] [CrossRef]
- Luchini, A.; Vitiello, G. Understanding the nano-bio interfaces: Lipid-coatings for inorganic nanoparticles as promising strategy for biomedical applications. Front. Chem. 2019, 7, 343. [Google Scholar] [CrossRef]
- Sivadasan, D.; Sultan, M.H.; Madkhali, O.; Almoshari, Y.; Thangavel, N. Polymeric lipid hybrid nanoparticles (plns) as emerging drug delivery platform—A comprehensive review of their properties, preparation methods, and therapeutic applications. Pharmaceutics 2021, 13, 1291. [Google Scholar] [CrossRef]
- Jordan, P.M.; Gerstmeier, J.; Pace, S.; Bilancia, R.; Rao, Z.; Börner, F.; Miek, L.; Gutiérrez-Gutiérrez, Ó.; Arakandy, V.; Rossi, A.; et al. Staphylococcus aureus-derived α-hemolysin evokes generation of specialized pro-resolving mediators promoting inflammation resolution. Cell Rep. 2020, 33, 108247. [Google Scholar] [CrossRef]
- Zhang, K.; Jordan, P.M.; Pace, S.; Hofstetter, R.K.; Werner, M.; Chen, X.; Werz, O. Modulation of Inflammation-Related Lipid Mediator Pathways by Celastrol During Human Macrophage Polarization. J. Inflamm. Res. 2022, 2022, 3285–3304. [Google Scholar] [CrossRef] [PubMed]
- Cinar, G.; Solomun, J.I.; Mapfumo, P.; Traeger, A.; Nischang, I. Nanoparticle sizing in the field of nanomedicine: Power of an analytical ultracentrifuge. Anal. Chim. Acta 2022, 1205, 339741. [Google Scholar] [CrossRef] [PubMed]











| Sample | dH (nm) ± SD | PDI ± SD | ζ (mV) ± SD | Yield ± SD (%) n = 4 | LC (%) ± SD | dN (nm) |
|---|---|---|---|---|---|---|
| HNP-NH2 | 147 ± 4 | 0.14 ± 0.05 | −20 ± 6 | 47 ± 8 | 0.06 ± 0.00 | 87 |
| HNP-COOH | 150 ± 20 | 0.15 ± 0.10 | −35 ± 12 | 67 ± 13 | 0.07 ± 0.01 | 90 |
| HNP-RGD | 150 ± 20 | 0.08 ± 0.01 | −35 ± 5 | 72 ± 7 | 0.06 ± 0.01 | 98 |
| HNP-cRGD | 137 ± 3 | 0.07 ± 0.02 | −29 ± 5 | 41 ± 11 | 0.07 ± 0.01 | 96 |
| PEG-PLGA | 122 ± 4 | 0.07 ± 0.01 | −14 ± 15 | 64 ± 9 | 0.05 ± 0.00 | 72 |
| Sample | dH (nm) ± SD | PDI ± SD | ζ (mV) ± SD | LCBRP-201 (%) ± SD | LCNLO (%) ± SD | dN (nm) |
|---|---|---|---|---|---|---|
| s-HNP-COOH | 153 ± 10 | 0.32 ± 0.04 | −40 ± 6 | 1.24 ± 0.22 | 0.07 ± 0.01 | 94 |
| s-HNP-RGD | 143 ± 5 | 0.27 ± 0.05 | −36 ± 4 | 1.00 ± 0.07 | 0.06 ± 0.01 | 99 |
| s-HNP-COOH/RGD (1:1) | 139 ± 4 | 0.24 ± 0.04 | −41 ± 7 | 1.07 ± 0.35 | 0.07 ± 0.01 | 89 |
| s-HNP-COOH/RGD (2:1) | 142 ± 5 | 0.25 ± 0.03 | −35 ± 2 | 1.15 ± 0.21 | 0.07 ± 0.01 | 77 |
| s-PEG-PLGA | 137 ± 33 | 0.23 ± 0.09 | −24 ± 6 | 1.33 ± 0.28 | 0.06 ± 0.01 | 84 |
| l-HNP-COOH | 249 ± 19 | 0.29 ± 0.03 | −41 ± 0 | 1.37 ± 0.29 | 0.08 ± 0.00 | 150 |
| l-HNP-RGD | 234 ± 16 | 0.25 ± 0.05 | −34 ± 7 | 1.38 ± 0.40 | 0.08 ± 0.00 | 162 |
| l-HNP-COOH/RGD (1:1) | 256 ± 25 | 0.28 ± 0.03 | −35 ± 2 | 1.70 ± 0.11 | 0.08 ± 0.00 | 194 |
| l-HNP-COOH/RGD (2:1) | 252 ± 19 | 0.27 ± 0.32 | −35 ± 7 | 1.38 ± 0.22 | 0.07 ± 0.01 | 140 |
| PEG-PLGA | 174 ± 6 | 0.21 ± 0.04 | −24 ± 0 | 2.89 ± 0.33 | 0.11 ± 0.01 | 143 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, J.; Klepsch, L.C.; Dahlke, P.; Tsarenko, E.; Vollrath, A.; Pretzel, D.; Jordan, P.M.; Rezaei, K.; Czaplewska, J.A.; Stumpf, S.; et al. PEG–Lipid–PLGA Hybrid Particles for Targeted Delivery of Anti-Inflammatory Drugs. Pharmaceutics 2024, 16, 187. https://doi.org/10.3390/pharmaceutics16020187
Ismail J, Klepsch LC, Dahlke P, Tsarenko E, Vollrath A, Pretzel D, Jordan PM, Rezaei K, Czaplewska JA, Stumpf S, et al. PEG–Lipid–PLGA Hybrid Particles for Targeted Delivery of Anti-Inflammatory Drugs. Pharmaceutics. 2024; 16(2):187. https://doi.org/10.3390/pharmaceutics16020187
Chicago/Turabian StyleIsmail, Jana, Lea C. Klepsch, Philipp Dahlke, Ekaterina Tsarenko, Antje Vollrath, David Pretzel, Paul M. Jordan, Kourosh Rezaei, Justyna A. Czaplewska, Steffi Stumpf, and et al. 2024. "PEG–Lipid–PLGA Hybrid Particles for Targeted Delivery of Anti-Inflammatory Drugs" Pharmaceutics 16, no. 2: 187. https://doi.org/10.3390/pharmaceutics16020187