Potentiation of Antibiotic Activity of Aztreonam against Metallo-β-Lactamase-Producing Multidrug-Resistant Pseudomonas aeruginosa by 3-O-Substituted Difluoroquercetin Derivatives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Docking Study
2.2. Chemicals and Reagents
2.3. Synthesis
2.4. Microorganisms
2.5. Assessment of Antibacterial Activity
2.6. Inhibition of Efflux Pumps: EtBr Accumulation
2.7. Inhibition of β-Lactamase
2.8. Antibiotics-Potentiation Activity
(in combination with ATM)/MIC (test compound alone)
2.9. Time–Kill Assay
2.10. Statistical Analysis
3. Results and Discussion
3.1. Clinical MBL-Producing P. aeruginosa Strains Showed High-Level Antibiotics Resistance
3.2. Efflux Pump and β-Lactamase Inhibitors Potentiated ATM against Highly Resistant MBL-Producing P. aeruginosa
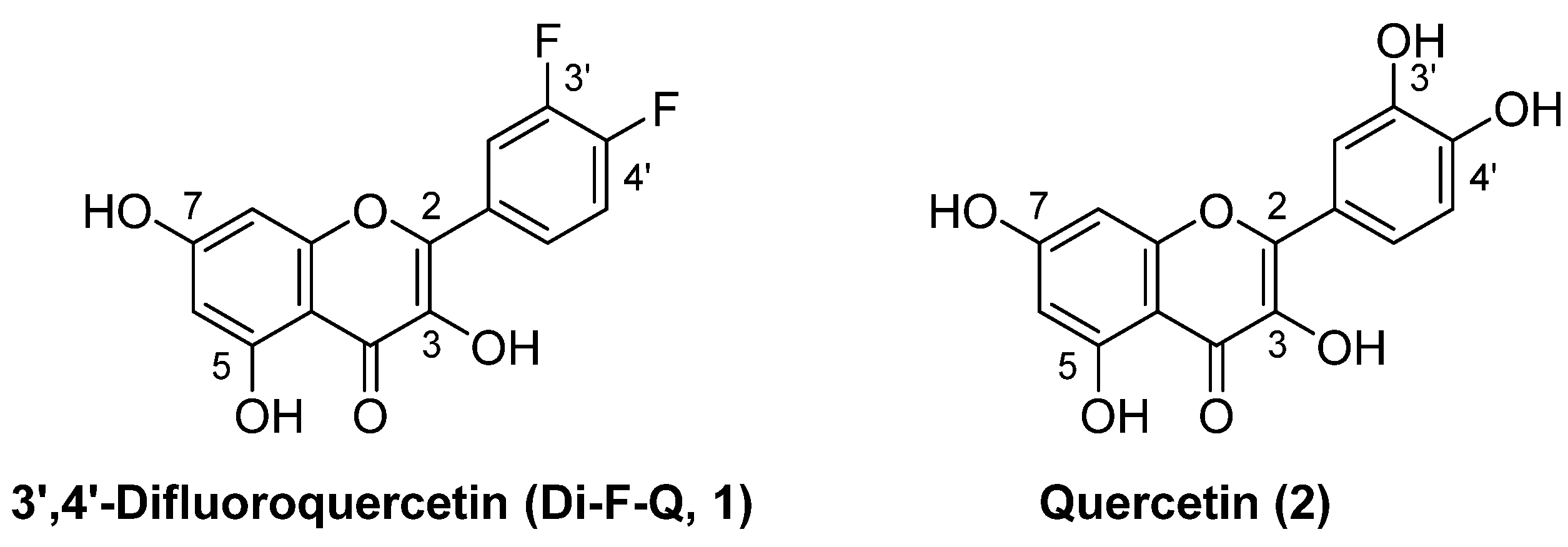
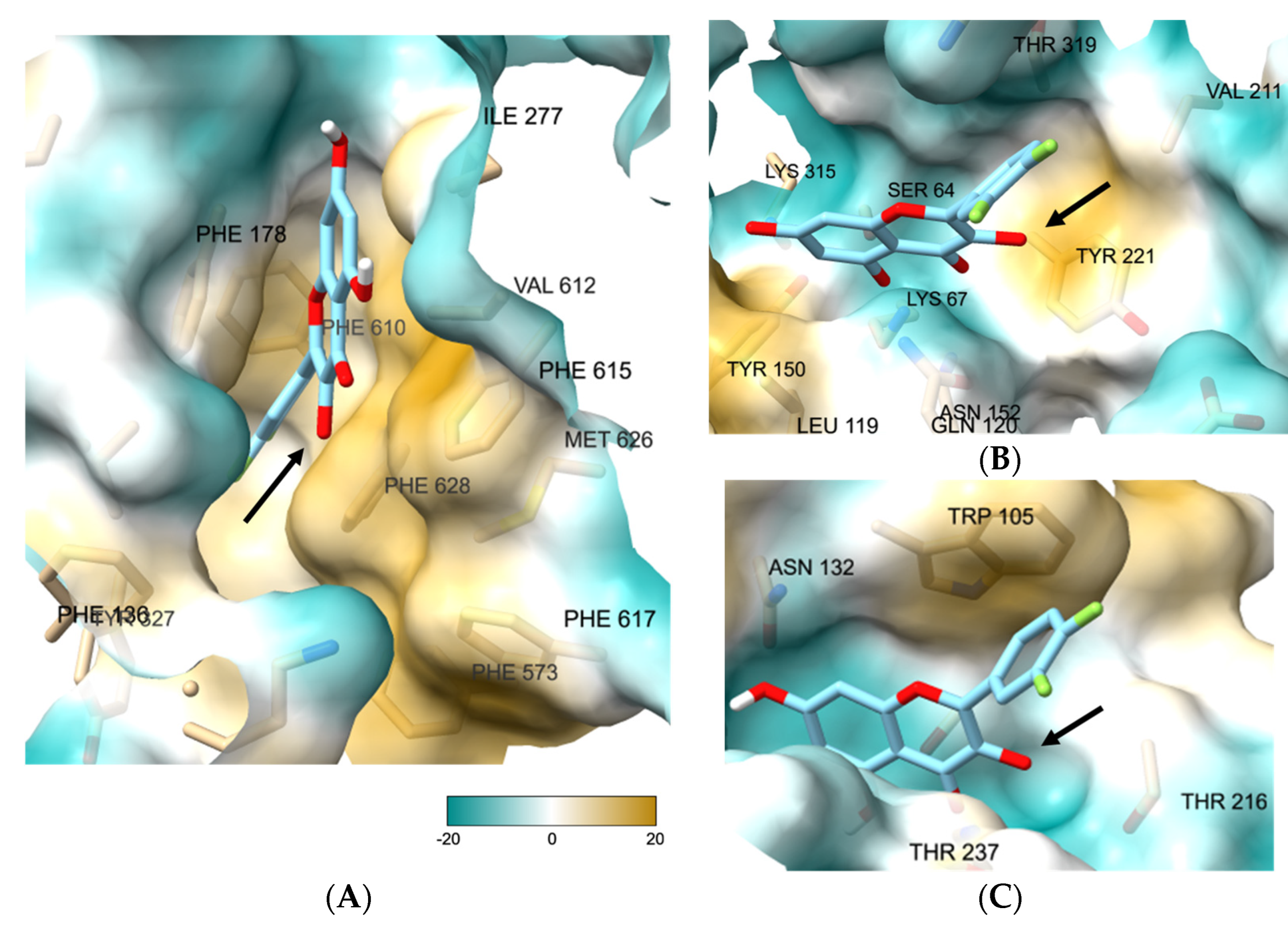
3.3. 3-O-Substituted di-F-Q (1) Showed Multitarget Inhibitory Activity against Efflux Pumps and β-Lactamases
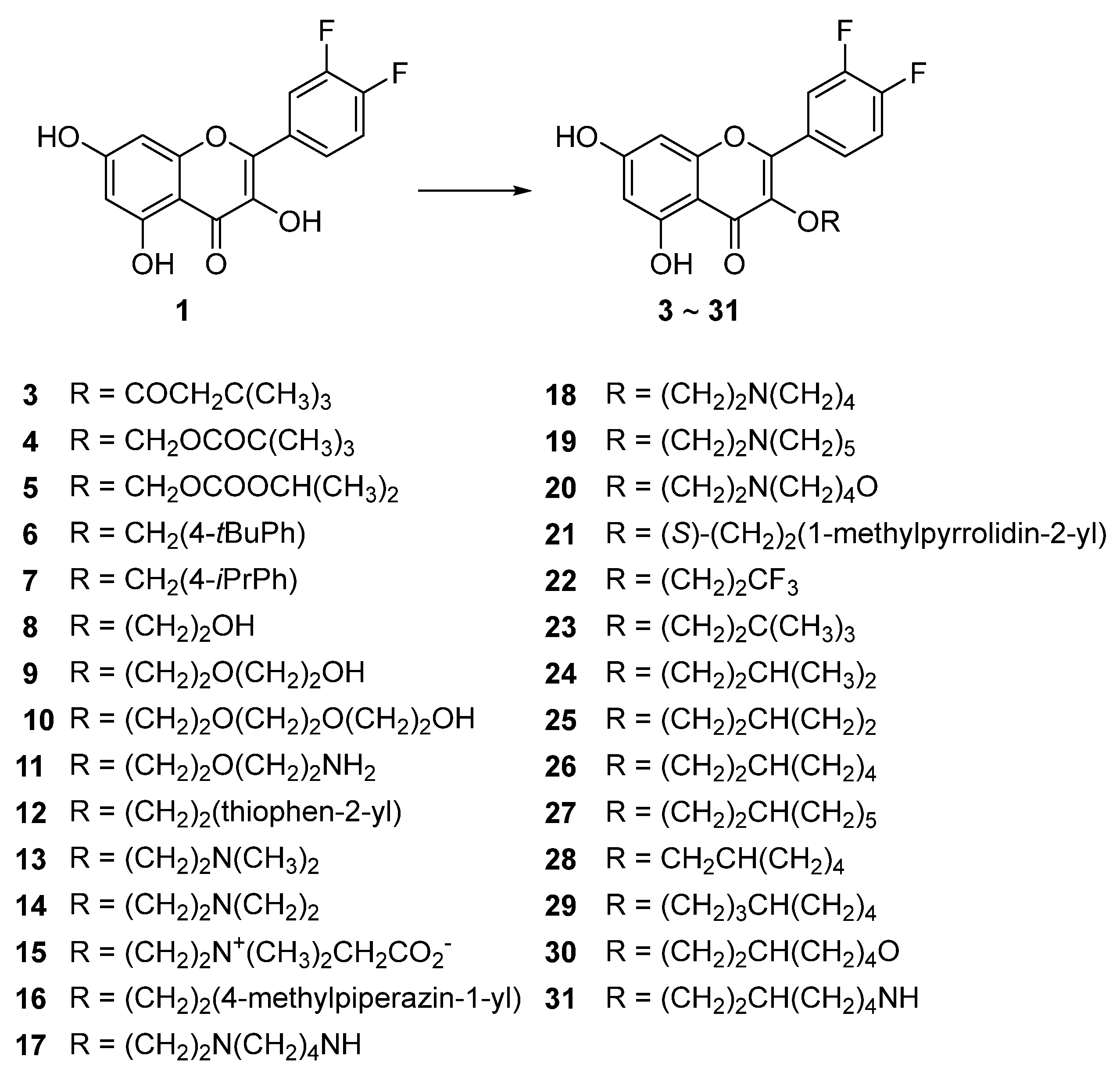
3.4. Syntheses of the Title Compounds, 3-O-Alkyl-di-F-Q Derivatives (3–31)
3.5. Compounds 23, 26, and 27 Showed Concentration-Dependent Efflux Pump Inhibitory Activity in MBL-Producing P. aeruginosa
3.6. Compounds 23, 26, and 27 Exhibited Broad-Spectrum β-Lactamase Inhibitory Activity
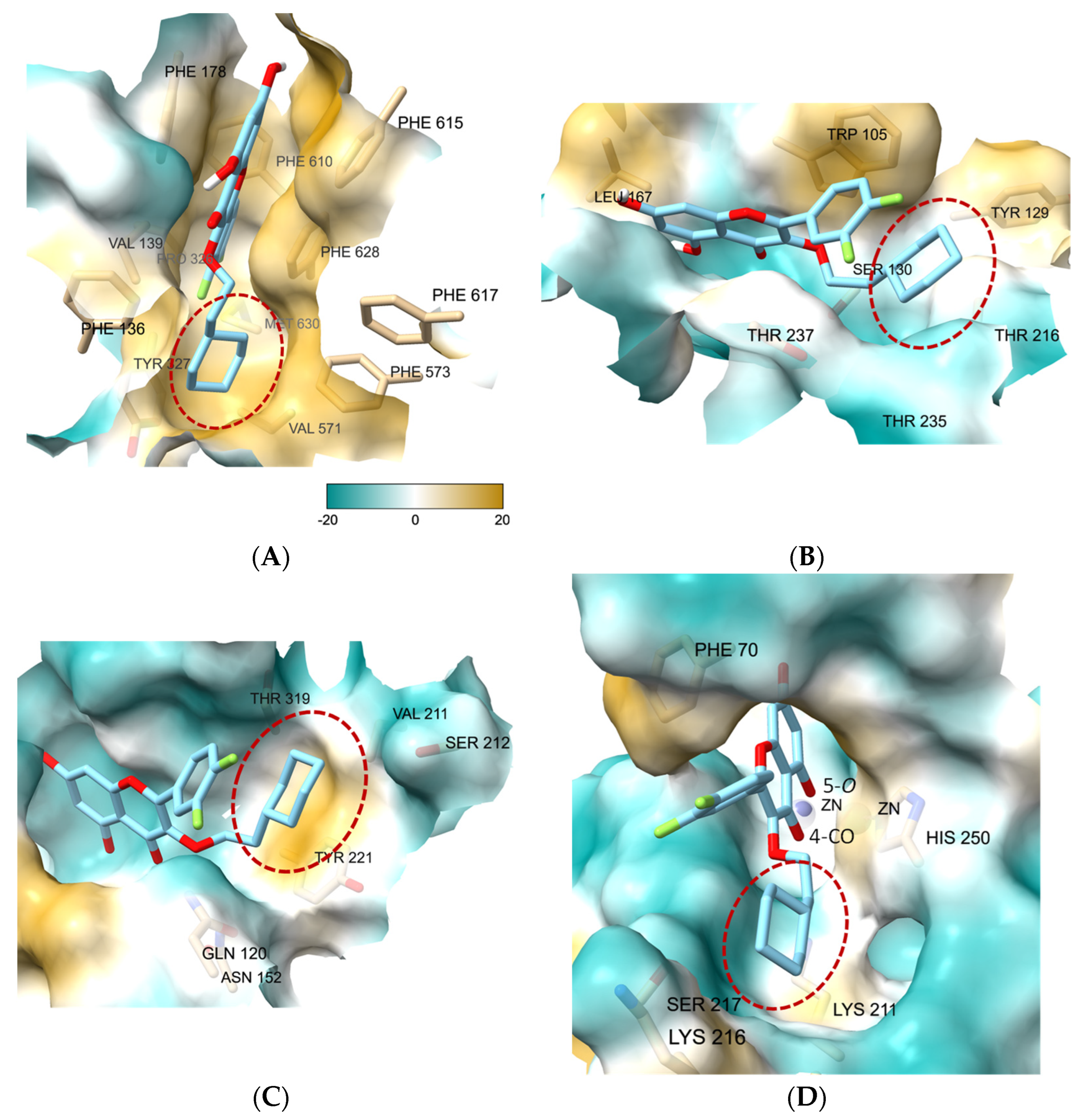
3.7. Molecular Docking Study of 27 Showed the Binding Role of Its 3-O-Substituent to the Efflux Pump and the Broad-Spectrum β-Lactamases
3.8. Antimicrobial Activity of ATM Was Significantly Increased by Combination with 27
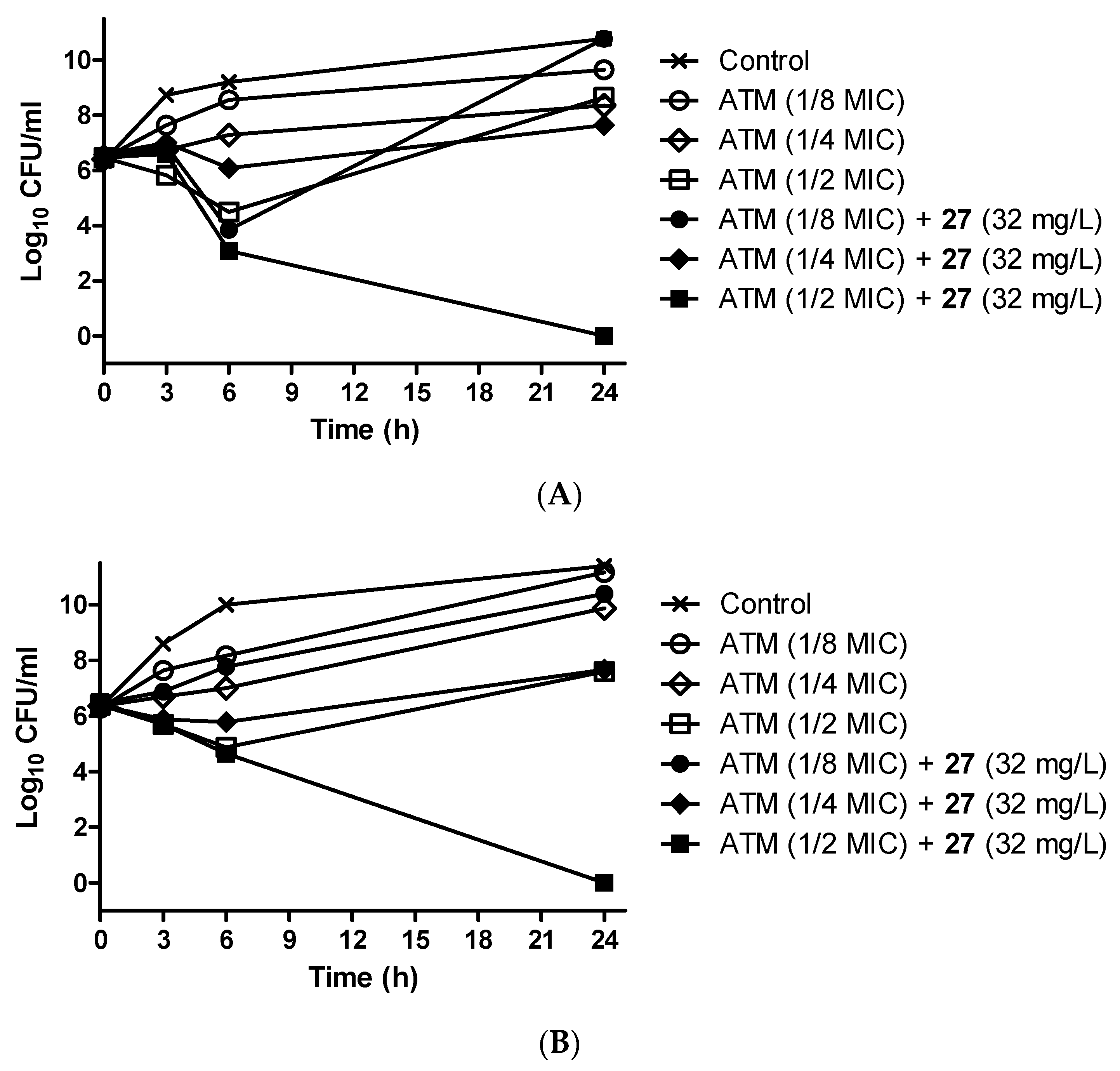
3.9. Antibiotic-Potentiating Activity of 27 Was Confirmed by Time–Kill Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Armstrong, T.; Fenn, S.J.; Hardie, K.R. JMM Profile: Carbapenems: A broad-spectrum antibiotic. J. Med. Microbiol. 2021, 70, 001462. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M. Antibiotics resistance: The last resort. Nature 2013, 499, 394–396. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem resistance: A review. Med. Sci. 2018, 6, 1. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T. Emerging carbapenemases: A global perspective. Int. J. Antimicrob. Agents 2010, 36, S8–S14. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Caliskan-Aydogan, O.; Alocilja, E.C. A review of carbapenem resistance in Enterobacterales and its detection techniques. Microorganisms 2023, 11, 1491. [Google Scholar] [CrossRef] [PubMed]
- Arer, V.; Kar, D. Biochemical exploration of β-lactamase inhibitors. Front. Genet. 2023, 13, 1060736. [Google Scholar] [CrossRef]
- Reddy, N.; Shungube, M.; Arvidsson, P.I.; Baijnath, S.; Kruger, H.G.; Govender, T.; Naicker, T. A 2018-2019 patent review of metallo beta-lactamase inhibitors. Expert Opin. Ther. Pat. 2020, 30, 541–555. [Google Scholar] [CrossRef]
- Mojica, M.F.; Bonomo, R.A.; Fast, W. B1-metallo-β-lactamases: Where do we stand? Curr. Drug Targets 2016, 17, 1029–1050. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Rubio-Aparicio, D.; Nelson, K.; Sun, D.; Tsivkovski, R.; Castanheira, M.; Lindley, J.; Loutit, J.; Dudley, M. In vitro activity of the ultrabroad-spectrum beta-lactamase inhibitor QPX7728 in combination with multiple beta-lactam antibiotics against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2021, 65, e00210-21. [Google Scholar] [CrossRef]
- Yasmin, M.; Fouts, D.E.; Jacobs, M.R.; Haydar, H.; Marshall, S.H.; White, R.; D’Souza, R.; Lodise, T.P.; Rhoads, D.D.; Hujer, A.M.; et al. Monitoring ceftazidime-avibactam and aztreonam concentrations in the treatment of a bloodstream infection caused by a multidrug-resistant Enterobacter sp. carrying both Klebsiella pneumoniae carbapenemase-4 and New Delhi metallo-β-lactamase-1. Clin. Infect. Dis. 2020, 71, 1095–1098. [Google Scholar] [CrossRef]
- Marshall, S.; Hujer, A.M.; Rojas, L.J.; Papp-Wallace, K.M.; Humphries, R.M.; Spellberg, B.; Hujer, K.M.; Marshall, E.K.; Rudin, S.D.; Perez, F.; et al. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob. Agents Chemother. 2017, 61, e02243-16. [Google Scholar] [CrossRef]
- Shaw, E.; Rombauts, A.; Tubau, F.; Padullés, A.; Càmara, J.; Lozano, T.; Cobo-Sacristán, S.; Sabe, N.; Grau, I.; Rigo-Bonnin, R.; et al. Clinical outcomes after combination treatment with ceftazidime/avibactam and aztreonam for NDM-1/OXA-48/CTX-M-15-producing Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 2018, 73, 1104–1106. [Google Scholar] [CrossRef]
- Alm, R.A.; Johnstone, M.R.; Lahiri, S.D. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: Role of a novel insertion in PBP3. J. Antimicrob. Chemother. 2015, 70, 1420–1428. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Rabine, S.; Hackel, M.; McLaughlin, R.E.; Biedenbach, D.J.; Bouchillon, S.K.; Sahm, D.F.; Bradford, P.A. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 1067–1078. [Google Scholar] [CrossRef]
- Periasamy, H.; Joshi, P.; Palwe, S.; Shrivastava, R.; Bhagwat, S.; Patel, M. High prevalence of Escherichia coli clinical isolates in India harbouring four amino acid inserts in PBP3 adversely impacting activity of aztreonam/avibactam. J. Antimicrob. Chemother. 2020, 75, 1650–1651. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nicolas, D.; Collet, L.; Bellais, S.; Cavallo, J.-D.; Nordmann, P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 2000, 44, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Iyobe, S.; Inoue, M.; Mitsuhashi, S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1991, 35, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Biedenbach, D.J.; Kazmierczak, K.; Bouchillon, S.K.; Sahm, D.F.; Bradford, P.A. In vitro activity of aztreonam–avibactam against a global collection of Gram-negative pathogens from 2012 and 2013. Antimicrob. Agents Chemother. 2015, 59, 4239–4248. [Google Scholar] [CrossRef] [PubMed]
- Mesaros, N.; Nordmann, P.; Plésiat, P.; Roussel-Delvallez, M.; van Eldere, J.; Glupczynski, Y.; van Laethem, Y.; Jacobs, F.; Lebecque, P.; Malfroot, A.; et al. Pseudomonas aeruginosa: Resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 2007, 13, 560–578. [Google Scholar] [CrossRef]
- Giamarellou, H. Prescribing guidelines for severe Pseudomonas infections. J. Antimicrob. Chemother. 2002, 49, 229–233. [Google Scholar] [CrossRef] [PubMed]
- McGowan, J.E. Resistance in nonfermenting gram-negative bacteria: Multidrug resistance to the maximum. Am. J. Med. 2006, 119, S29–S36. [Google Scholar] [CrossRef]
- Thomson, J.M.; Bonomo, R.A. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: Beta-lactams in peril! Curr. Opin. Microbiol. 2005, 8, 518–524. [Google Scholar] [CrossRef]
- Poole, K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004, 10, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Hocquet, D.; Nordmann, P.; El Garch, F.; Cabanne, L.; Plesiat, P. Involvement of the MexXY–OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 1347–1351. [Google Scholar] [CrossRef]
- Quale, J.; Bratu, S.; Gupta, J.; Landman, D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2006, 50, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, I.; Couetdic, G.; Clermont, O.; Brahimi, N.; Plesiat, P.; Bingen, E. In vivo selection of a target⁄efflux double mutant of Pseudomonas aeruginosa by ciprofloxacin therapy. J. Antimicrob. Chemother. 2001, 48, 553–555. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Sugimoto, Y.; Yoneyama, H.; Nakae, T. High-level fluoroquinolone resistance in Pseudomonas aeruginosa due to interplay of the MexAB–OprM efflux pump and the DNA gyrase mutation. Microbiol. Immunol. 2002, 46, 391–395. [Google Scholar] [CrossRef]
- Niga, T.; Ito, H.; Oyamada, Y.; Yamagishi, J.-I.; Kadono, M.; Nishino, T.; Gotoh, N.; Inoue, M. Cooperation between alteration of DNA gyrase genes and over-expression of MexB and MexX confers high-level fluoroquinolone resistance in Pseudomonas aeruginosa strains isolated from a patient who received a liver transplant followed by treatment with fluoroquinolones. Microbiol. Immunol. 2005, 49, 443–446. [Google Scholar]
- Poole, K.; Srikumar, R. Multidrug efflux in Pseudomonas aeruginosa: Components, mechanisms and clinical significance. Curr. Top. Med. Chem. 2001, 1, 59–71. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef]
- Ma, Z.; Xu, C.; Zhang, X.; Wang, D.; Pan, X.; Liu, H.; Zhu, G.; Bai, F.; Cheng, Z.; Wu, W.; et al. A MexR mutation which confers aztreonam resistance to Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 659808. [Google Scholar] [CrossRef]
- Jorth, P.; McLean, K.; Ratjen, A.; Secor, P.R.; Bautista, G.E.; Ravishankar, S.; Rezayat, A.; Garudathri, J.; Harrison, J.J.; Harwood, R.A.; et al. Evolved aztreonam resistance is multifactorial and can produce hypervirulence in Pseudomonas aeruginosa. mBio 2017, 8, e00517. [Google Scholar] [CrossRef]
- Waditzer, M.; Bucar, F. Flavonoids as inhibitors of bacterial efflux pumps. Molecules 2021, 26, 6904. [Google Scholar] [CrossRef]
- Dos Santos, J.F.S.; Tintino, S.R.; da Silva, A.R.P.; Dos S Barbosa, C.R.; Scherf, J.R.; de S Silveira, Z.; de Freitas, T.S.; de Lacerda Neto, L.J.; Barros, L.M.; de A Menezes, I.R.; et al. Enhancement of antibiotic activity by quercetin against Staphylococcus aureus efflux pumps. J. Bioenerg. Biomembr. 2021, 53, 157–167. [Google Scholar] [CrossRef]
- Kho, W.; Kim, M.K.; Jung, M.; Chong, Y.P.; Kim, Y.S.; Park, K.-H.; Chong, Y. Strain-specific anti-biofilm and antibiotic-potentiating activity of 3′,4′-difluoroquercetin. Sci. Rep. 2020, 10, 14162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, C.; Cheng, B.; Gao, L.; Qin, C.; Zhang, L.; Zhang, X.; Wang, J.; Wan, Y. Discovery of quercetin and its analogs as potent OXA-48 beta-lactamase inhibitors. Front. Pharmacol. 2022, 13, 926104. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Tripathi, A. Quercetin inhibits carbapenemase and efflux pump activities among carbapenem-resistant Gram-negative bacteria. APMIS 2020, 128, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Benin, B.M.; Hillyer, T.; Shin, W.S. Quercetin, a potential metallo-β-lactamase inhibitor for use in combination therapy against β-lactam antibiotic-resistant bacteria. Open J. Pathol. Toxicol. Res. 2021, 1, OJPTR.MS.ID.000503. [Google Scholar]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Hayashi, K.; Nagata, C.; Hoshino, K.; Onodera, Y.; Nishino, K.; Yamaguchi, A. Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013, 500, 102–107. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, S.; Balius, T.E.; Singh, I.; Levit, A.; Moroz, Y.S.; O’Meara, M.J.; Che, T.; Algaa, E.; Tolmachova, K.; et al. Ultra-large library docking for discovering new chemotypes. Nature 2019, 566, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Hecker, S.J.; Reddy, K.R.; Lomovskaya, O.; Griffith, D.C.; Rubio-Aparicio, D.; Nelson, K.; Tsivkovski, R.; Sun, D.; Sabet, M.; Tarazi, Z.; et al. Discovery of cyclic boronic acid QPX7728, an ultrabroad-spectrum inhibitor of serine and metallo-β-lactamases. J. Med. Chem. 2020, 63, 7491–7507. [Google Scholar] [CrossRef] [PubMed]
- Pedretti, A.; Mazzolari, A.; Gervasoni, S.; Fumagalli, L.; Vistoli, G. The Vega suite of programs: A versatile platform for cheminformatics and drug design projects. Bioinformatics 2021, 37, 1174–1175. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Cho, S.Y.; Kim, M.K.; Mok, H.; Choo, H.; Chong, Y. Separation of quercetin’s biological activity from its oxidative property through bioisosteric replacement of the catecholic hydroxyl groups with fluorine atoms. J. Agric. Food Chem. 2012, 60, 6499–6506. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI M100-ED33:2023; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2023. [Google Scholar]
- Paixão, L.; Rodrigues, L.; Couto, I.; Martins, M.; Fernandes, P.; de Carvalho, C.C.; Monteiro, G.A.; Sansonetty, F.; Amaral, L.; Viveiros, M. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 2009, 3, 18. [Google Scholar] [CrossRef]
- Pemberton, O.A.; Jaishankar, P.; Akhtar, A.; Adams, J.L.; Shaw, L.N.; Renslo, A.R.; Chen, Y. Heteroaryl phosphonates as noncovalent inhibitors of both serine and metallocarbapenemases. J. Med. Chem. 2019, 62, 8480–8496. [Google Scholar] [CrossRef]
- Liu, B.; Trout, R.E.L.; Chu, G.-H.; McGarry, D.; Jackson, R.W.; Hamrick, J.C.; Daigle, D.M.; Cusick, S.M.; Pozzi, C.; De Luca, F.; et al. Discovery of taniborbactam (VNRX-5133): A broad-spectrum serine- and metallo-β-lactamase inhibitor for carbapenem-resistant bacterial infections. J. Med. Chem. 2020, 63, 2789–2801. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Fisher, J.F. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu. Rev. Microbiol. 2011, 65, 455–478. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Kim, Y.A.; Kim, J.; Jeong, S.H.; Shin, J.H.; Shin, K.S.; Shin, J.H.; Kim, Y.R.; Kim, H.S.; Uh, Y.; et al. Emergence of NDM-1-producing Pseudomonas aeruginosa sequence type 773 clone: Shift of carbapenemases molecular epidemiology and spread of 16S rRNA methylase genes in Korea. Ann. Lab. Med. 2023, 43, 196–199. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant, and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P.; et al. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: A retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin. Infect. Dis. 2018, 67, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Tripathi, A. Demonstration of bactericidal and synergistic activity of quercetin with meropenem among pathogenic carbapenem-resistant Escherichia coli and Klebsiella pneumoniae. Microb. Pathog. 2020, 143, 104120. [Google Scholar] [CrossRef]
- Sariyer, E.; Saral, A. Evaluation of quercetin as a potential β-lactamase CTX-M-15 inhibitor via molecular docking, dynamic simulations, and MMGBSA. Turk. J. Chem. 2021, 45, 1045–1056. [Google Scholar] [CrossRef]
- Sjuts, H.; Vargiu, A.V.; Kwasny, S.M.; Nguyen, S.T.; Kim, H.-S.; Ding, X.; Ornik, A.R.; Ruggerone, P.; Bowlin, T.L.; Nikaido, H.; et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, 3509–3514. [Google Scholar] [CrossRef]
- Viveiros, M.; Martins, A.; Paixão, L.; Rodrigues, L.; Martins, M.; Couto, I.; Fähnrich, E.; Kern, W.V. Amaral, Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by automated ethidium bromide method. Int. J. Antimicrob. Agents 2008, 31, 458–462. [Google Scholar] [CrossRef]
- Barnes, M.D.; Winkler, M.L.; Taracila, M.A.; Page, M.G.; Desarbre, E.; Kreiswirth, B.N.; Shields, R.H.; Nguyen, M.-H.; Clancy, C.; Spellberg, B.; et al. Klebsiella pneumoniae Carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: Unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 2017, 8, e00528-17. [Google Scholar] [CrossRef] [PubMed]
- Lohans, C.T.; Brem, J.; Schofield, C.J. New Delhi metallo-β-lactamase 1 catalyzes avibactam and aztreonam hydrolysis. Antimicrob. Agents Chemother. 2017, 61, e01224-17. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef] [PubMed]

| Isolates, MBLs Produced | MIC Value (mg/L) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP-TZB | CAZ-AVI | CAZ | FEP | ATM | IPM | MEM | AMK | GEN | CST | CIP | |
| PA-001, blaNDM | >64/4 | >8/4 | >16 | >16 | 32 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-007, blaNDM | >64/4 | >8/4 | >16 | 16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-008, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-011, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-012, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | ≤4 | 8 | ≤1 | >2 |
| PA-017, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-019, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-020, blaNDM | >64/4 | >8/4 | >16 | 16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-023, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-025, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-028, blaNDM | >64/4 | >8/4 | >16 | >16 | 8 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-034, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-036, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-038, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-039, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | ≤4 | ≤2 | ≤1 | >2 |
| PA-044, blaNDM | >64/4 | >8/4 | >16 | >16 | 16 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-002, blaIMP | >64/4 | >8/4 | >16 | >16 | 64 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-016, blaIMP | >64/4 | >8/4 | >16 | >16 | 64 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-027, blaIMP | 64/4 | 8/4 | >16 | >16 | 32 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-029, blaIMP | >64/4 | >8/4 | >16 | >16 | 64 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| PA-030, blaIMP | >64/4 | >8/4 | >16 | >16 | 64 | >8 | >32 | >32 | >8 | ≤1 | >2 |
| Isolates, MBLs Produced | MIC Value (mg/L) 2 | ||||||
|---|---|---|---|---|---|---|---|
| ATM-ARMA | |||||||
| ATM Alone | ATM-AVI | ATM-CZA | ATM-CCCP | ATM-PAβN | ATM-1 | ATM-CCCP-AVI | |
| PA-025, blaNDM | 16 | 16 | 16 | – | – | 8 (8) | – |
| PA-038, blaNDM | 16 | 16 | 16 | 4 (256) | 4 (128) | 8 (8) | – |
| PA-002, blaIMP | 64 | 64 | 16 (128) | 16 (256) | 32 (128) | 16 (16) | 4 (256/256) |
| PA-003, blaVIM | 32 | 16 (128) | 16 (128) | 8 (256) | 8 (128) | 16 (16) | 4 (256/256) |
| Compound | β-Lactamases, IC50 (μM) | Compound | β-Lactamases, IC50 (μM) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AmpC | KPC-2 | OXA-48 | VIM-2 | NDM-1 | AmpC | KPC-2 | OXA-48 | VIM-2 | NDM-1 | ||
| 1 | >50 | >50 | >100 | >100 | >50 | 17 | >50 | >50 | >100 | >100 | >50 |
| 3 | 19.2 | 22.2 | 21.2 | 20.3 | 24.6 | 18 | >50 | >50 | >100 | >100 | >50 |
| 4 | 25.4 | 33.2 | 28.7 | 19.3 | 15.7 | 19 | >50 | >50 | >100 | >100 | >50 |
| 5 | >50 | >50 | 42.7 | >100 | >50 | 20 | >50 | >50 | >100 | >100 | >50 |
| 6 | >50 | >50 | 37.8 | >100 | 14.1 | 21 | >50 | >50 | >100 | >100 | >50 |
| 7 | >50 | 21.5 | 6.0 | 13.1 | 16.7 | 22 | >50 | >50 | 39.8 | >100 | >50 |
| 8 | >50 | >50 | >100 | >100 | >50 | 23 | 18.9 | 17.3 | 14.8 | 11.9 | 11.4 |
| 9 | >50 | >50 | >100 | >100 | >50 | 24 | >50 | 39.4 | 19.1 | 42.1 | >50 |
| 10 | >50 | >50 | >100 | >100 | >50 | 25 | >50 | 22.8 | 11.4 | 13.9 | 22.7 |
| 11 | >50 | >50 | >100 | >100 | >50 | 26 | 21.0 | 11.5 | 8.0 | 9.0 | 15.9 |
| 12 | >50 | >50 | >100 | >100 | >50 | 27 | 15.3 | 11.3 | 6.3 | 4.4 | 10.1 |
| 13 | >50 | >50 | >100 | >100 | >50 | 28 | >50 | 40.8 | 9.2 | 7.8 | 21.8 |
| 14 | >50 | >50 | >100 | >100 | >50 | 29 | >50 | 28.3 | 5.5 | 5.1 | 26.7 |
| 15 | >50 | >50 | >100 | >100 | >50 | 30 | >50 | >50 | 35.9 | >100 | >50 |
| 16 | >50 | >50 | >100 | >100 | >50 | 31 | >50 | >50 | >100 | >100 | >50 |
| Isolates, MBLs Produced | Effects of Combination on the MIC of ATM | |||||
|---|---|---|---|---|---|---|
| ATM (Alone) | ATM-27 | ATM-AVI | ATM-CCCP | ATM-CCCP-AVI | ||
| MIC 1 | FD 2 (FICI) 3 | MIC 1 | MIC 1 | MIC 1 | ||
| PA-001, blaNDM | 32 | 16 (8) | 2 (A) | 16 (128) | 16 (32) | 16 (16/64) |
| PA-007, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 8 (64) | 8 (64/– 4) |
| PA-008, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 8 (64) | 8 (64/– 4) |
| PA-011, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 8 (64) | 8 (64/– 4) |
| PA-012, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 8 (64) | 8 (64/– 4) |
| PA-017, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-019, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-020, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-023, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-025, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-028, blaNDM | 8 | 1 (32) | 8 (S) | 8 4 | 8 4 | 8 4 |
| PA-034, blaNDM | 16 | 4 (16) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-036, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-038, blaNDM | 16 | 4 (8) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-039, blaNDM | 16 | 4 (8) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-044, blaNDM | 16 | 4 (32) | 4 (S) | 16 4 | 16 4 | 16 4 |
| PA-002, blaIMP | 64 | 8 (32) | 8 (S) | 64 4 | 64 4 | 64 4 |
| PA-016, blaIMP | 64 | 8 (16) | 8 (S) | 64 4 | 64 4 | 64 4 |
| PA-027, blaIMP | 32 | 8 (8) | 4 (S) | 32 4 | 32 4 | 32 4 |
| PA-029, blaIMP | 64 | 16 (16) | 4 (S) | 64 4 | 64 4 | 64 4 |
| PA-030, blaIMP | 64 | 32 (8) | 2 (A) | 64 4 | 64 4 | 64 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Lee, T.; Kim, M.K.; Ahn, J.H.; Jeong, S.; Park, K.-H.; Chong, Y. Potentiation of Antibiotic Activity of Aztreonam against Metallo-β-Lactamase-Producing Multidrug-Resistant Pseudomonas aeruginosa by 3-O-Substituted Difluoroquercetin Derivatives. Pharmaceutics 2024, 16, 185. https://doi.org/10.3390/pharmaceutics16020185
Lee S, Lee T, Kim MK, Ahn JH, Jeong S, Park K-H, Chong Y. Potentiation of Antibiotic Activity of Aztreonam against Metallo-β-Lactamase-Producing Multidrug-Resistant Pseudomonas aeruginosa by 3-O-Substituted Difluoroquercetin Derivatives. Pharmaceutics. 2024; 16(2):185. https://doi.org/10.3390/pharmaceutics16020185
Chicago/Turabian StyleLee, Seongyeon, Taegum Lee, Mi Kyoung Kim, Joong Hoon Ahn, Seri Jeong, Ki-Ho Park, and Youhoon Chong. 2024. "Potentiation of Antibiotic Activity of Aztreonam against Metallo-β-Lactamase-Producing Multidrug-Resistant Pseudomonas aeruginosa by 3-O-Substituted Difluoroquercetin Derivatives" Pharmaceutics 16, no. 2: 185. https://doi.org/10.3390/pharmaceutics16020185





