The Role of Bacteriophages in the Gut Microbiota: Implications for Human Health
Abstract
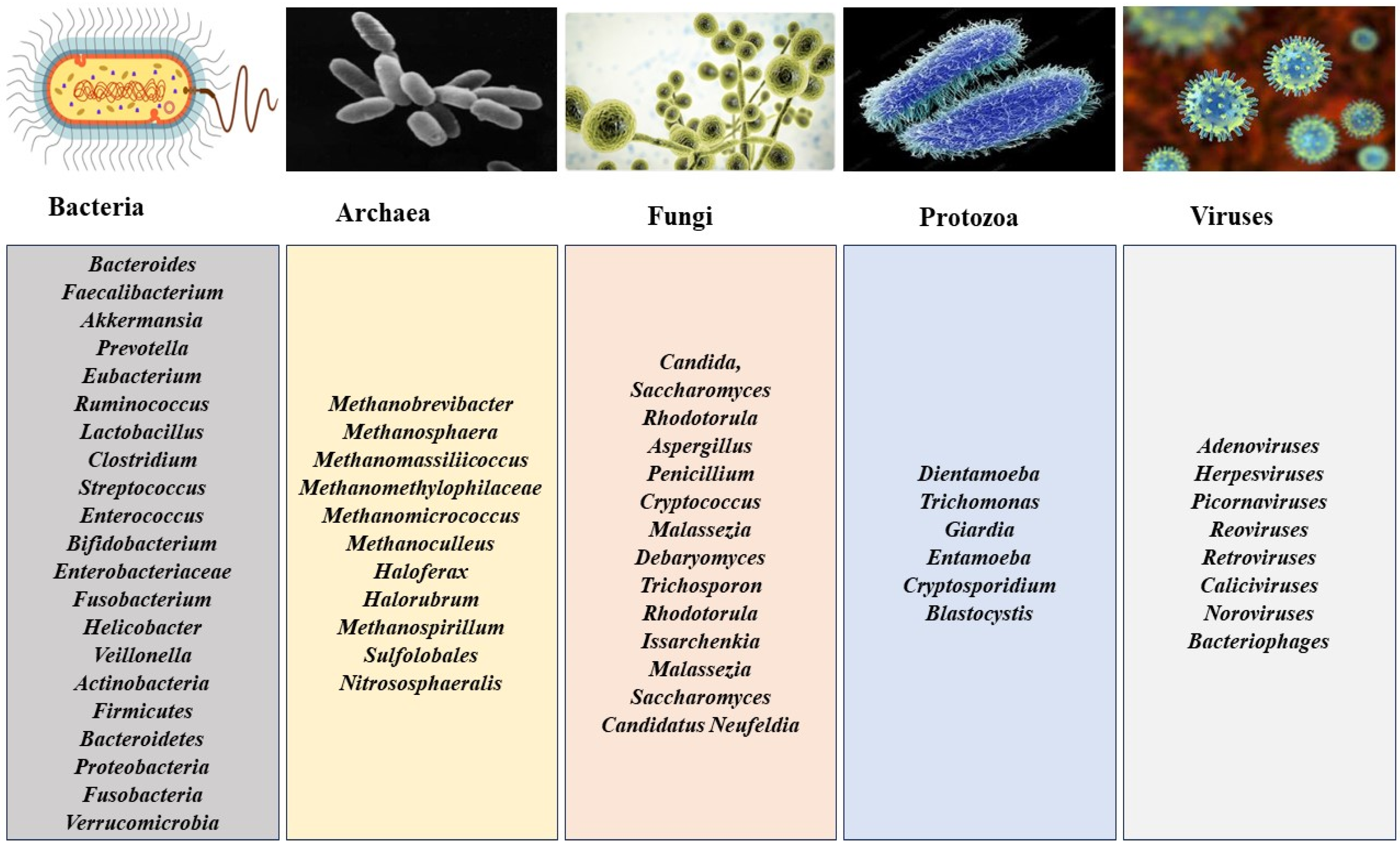
:1. Introduction
2. Gut Microbiota and Composition
3. Functions of the Gut Microbiota
3.1. Effects of Gut Microbiota on Host Metabolism
3.2. Immunomodulatory Potential of Gut Microbiota
3.3. Bioprotection against Invading Pathogens
3.4. Effect on Xenobiotics and Drug Metabolism
3.5. Protection of Mucosal Integrity
4. Bacteriophages: Classification, Life Cycle, and General Mechanism of Action
5. Impact and Roles in Maintaining the Balance of the Gut Microbiota
6. Potential Therapeutic Applications
6.1. Therapeutic Applications of Bacteriophages against Pathogenic Vibrio spp.
6.2. Therapeutic Applications of Bacteriophages against Pathogenic Escherichia coli
6.3. Therapeutic Applications of Bacteriophages against Pathogenic Clostridioides Difficile
6.4. Therapeutic Applications of Bacteriophages against Pathogenic Salmonella spp.
6.5. Therapeutic Applications of Bacteriophages against Pathogenic Fusobacterium nucleatum
6.6. Therapeutic Applications of Bacteriophages against Pathogenic Shigella spp.
6.7. Therapeutic Applications of Bacteriophages against Pathogenic Klebsiella pneumoniae
6.8. Therapeutic Applications of Bacteriophages against Pathogenic Listeria monocytogenes
6.9. Therapeutic Applications of Bacteriophages against Pathogenic Ruminococcus gnavus
6.10. Therapeutic Applications of Bacteriophages against Pathogenic Campylobacter spp.
6.11. General
| Target Pathogen | Phage Therapy | Disease | Study Type, Model | Reports | References |
|---|---|---|---|---|---|
| Vibrio spp. | Cocktail of five (5) lytic Vibrio phages | Fighting V. cholerae infection | In vivo, rabbits | Oral administration of cocktails before infection resulted in prophylactic effects. The phage cocktail significantly reduced bacterial load 6 and 12 h after the challenge. | Jaiswal et al. [109] |
| Oral phage cocktail therapy | V. cholerae infection | In vivo, mice | Phage cocktail (108 PFU/mL) given once daily significantly reduced bacterial load. | Jaiswal et al. [110] | |
| Bacteriophage pVp-1 | Multiple antibiotic-resistant V. parahaemolyticus implicated in gastroenteritis | In vivo, mice | Protection from infection and death 1 h after inoculation with V. parahaemolyticus. | Jun et al. [111] | |
| Cocktail of three (3) lytic (virulent) phages—ICP1, ICP2, and ICP3 | Cholera pathogenesis/cholera-like diarrhea | In vivo, infant mice and rabbits | Effective at preventing mouse small intestinal colonization. Prophylaxis against the onset of cholera-like diarrhea was achieved after oral administration of the phages up to 24 h before V. cholera infestation. | Yen et al. [112] | |
| Escherichia coli | Cocktail of three (3) bacteriophages from Siphoviridae and Podoviridae | E. coli implicated in gastrointestinal diseases | In vitro | Phage cocktail exhibited broad spectrum and strong lytic activity against E. coli isolates. | Nasr-Eldin et al. [113] |
| Cocktail of lytic phages specific against E. coli | Gut pathogenic E. coli | In vivo, mice | Suppression of E. coli was observed 5–10 d after phage therapy. | Abdulamir et al. [114] | |
| Lytic Myoviridae phage phPE42 | Extensively drug-resistant (XDR) E. coli implicated in foodborne infections | In vivo, rats | Effective eradication of XDR E. coli was observed in animal feces. | Abdelaziz et al. [115] | |
| T4-like phages | Childhood diarrhea-associated E. coli isolates | In vitro, cultures | T4-like phages combined in a cocktail resulted in increased bacterial lysis. | Bourdin et al. [116] | |
| Phage-based probiotic dietary supplement consisting of 7 bacteriophage strains | Traveler’s diarrhea (TD) caused by E. coli, S. flexneri, S. sonnei, S. enterica, L. monocytogenes, S. aureus | Clinical study, in vivo, humans and mice | Prophylactic effect against TD. | Aleshkin et al. [117] | |
| Specific bacteriophage | Enteropathogenic E. coli (EPEC) | In vivo, mice | A single dose of the phage rendered a protective effect on the bacteria throughout the study. | Vahedi et al. [118] | |
| T4-like coliphages | Acute bacterial diarrhea | Clinical trial, humans | Failure to improve diarrhea condition, possibly due to insufficient phage concentration. | Sarker et al. [119] | |
| Virulent bacteriophages targeting prototype of the Adherent Invasive E. coli (AIEC) strain LF82 | Crohn’s disease (CD) | Ex vivo, in vivo, murine and human intestinal samples | Three virulent bacteriophage cocktails were active against the AIEC strain LF82. A single dose of the cocktail reduced colitis symptoms in mice colonized with AIEC. | Galtier et al. [120] | |
| Bacteriophage cocktail Ec17B153DK1 vs. the broad-spectrumantibiotic ciprofloxacin | E. coli infecting the gut environment | In vitro, simulated small intestine system | The cocktail was effective in reducing E. coli in simulated gut conditions. No impact on commensal, non-targeted bacteria. | Cieplak et al. [121] | |
| Commercial cocktail of E. coli-targeting bacteriophages (PreforPro®) containing four phages (LH01-Myoviridae, LL5-Siphoviridae, T4D-Myoviridae, and LL12-Myoviridae) | Effect on gut microbiota during GI distress and markers of intestinal and systemic inflammation | Clinical trial, humans | The potential of bacteriophages to selectively reduce target organisms without causing dysbiosis | Gindin et al. [122] | |
| Supplemental bacteriophages (PreforPro®) | Enhance the effects of a common probiotic, B. animalis subsp. Lactis (B. lactis) on GI health | Clinical study, humans | Improvements in GI inflammation and colon pain in individuals consuming B. lactis with PreforPro®. | Grubb et al. [123] | |
| Suppository containing probiotic strains of Lactobacillus spp. and bacteriophages specific for pathogenic E. coli | Diarrhea | In vivo, calves | Probiotic-phage suppositories reduced the duration of diarrhea in calves. The complete stopping of diarrhea was observed 24–48 h after use. | Alomari et al. [124] | |
| Lytic phages (T4, F1, B40-8, and VD13 phages) | Effect on mice gut colonized with human commensal bacteria | In vivo, gnotobiotic mice | Targeted lysis of susceptible gut bacteria. Modulation of non-targeted bacteria through interbacterial interactions. | Hsu et al. [95] | |
| Genetically engineered temperate phages | Shiga-toxin (Stx)-producing E. coli colonizing the mammalian gut | In vivo, mice | Significant repression of fecal Stx concentrations. Suppression of virulence factors in gut bacteria. | Hsu et al. [125] | |
| Phage PDX, a member of the Myoviridae family | Diarrheagenic enteroaggregative E. coli (EAEC) | In vitro, in vivo, cultures and mice | Bacteriolytic activity of EAEC isolates (EN1E-0007) in vitro and in vivo. No dysbiosis was observed in the anaerobic culture. | Cepko et al. [126] | |
| Phage ES17, a Podoviridae phage | Extraintestinal pathogenic E. coli (ExPEC) in the intestine | In vivo, mice | Selective elimination of invasive pathobiont species from mucosal surfaces in the intestinal tract. | Green et al. [128] | |
| Clostridium difficile | Six (6) myoviruses and one (1) siphovirus | C. difficile infection (CDI) | In vitro, in vivo, hamsters | Specific phage combinations resulted in total lysis of C. difficile in vitro. Prevention of resistance. In vivo, the evaluation revealed a reduction in C. difficile colonization 36 h post-infection. | Nale et al. [129] |
| Recombinant bacteriophage | C. difficile infection | In vitro, in vivo, cultures and mice | Targeting and killing of C. difficile. | Selle et al. [130] | |
| Salmonella spp. | Phage SE20 (Podoviridae) | S. enterica serotype Enteritidis | In vitro, in vivo, mice | Oral administration of a single dose of bacteriophage protected against salmonellosis and treatment of salmonellosis. Animals developed hepatomegaly and splenomegaly as side effects but had no gastrointestinal complications with the phage therapy. | Dallal et al. [131] |
| Bacteriophage cocktail (foodborne outbreak pill (FOP) targeting E. coli O157:H7, L. monocytogenes, and Salmonella) | Salmonella infection | In vitro | Simulator of the Human Intestinal Microbial Ecosystem (SHIME). | Moye et al., 2019 [133] | |
| Phage cocktail | Salmonella colonization in experimentally challenged birds | In vivo, birds | Phage treatment effectively reduced Salmonella colonization and enhanced growth performance weight gains in challenged birds. | Thanki et al. [132] | |
| Myoviruses and a siphovirus | Salmonella infection gastrointestinal enteritis | In vitro, in vivo, swine, birds, cultures | Phage cocktail (STW-77 and SEW-109) had the most lysing efficacy on the swine and bird models. Some phages from the cocktail could lyse resistant strains of the organism. | Nale et al. [134] | |
| Salmonella phages (vB_SenS_KP001, vB_SenS_KP005, and vB_SenS_WP110) | Salmonella colonization in the gastrointestinal tract of broilers | In vivo, broilers | The phage cocktail reduced Salmonella colonization in broilers’ gastrointestinal tracts from over 70% to 0% 4 d post-treatment. | Pelyuntha et al. [135] | |
| Fusobacterium nucleatum | Irinotecan-loaded dextran nanoparticles covalently linked to azide-modified phages. | Colorectal cancer (CRC) | In vivo, mice | Phage administration inhibited the growth of F. nucleatum. It significantly boosted the effectiveness of first-line chemotherapy treatments for CRC. | Zheng et al. [136] |
| F. nucleatum (Fn)-binding M13-phage-loaded silver nanoparticles (AgNPs) | Symbiotic F. nucleatum in the gut selectively increases immunosuppressive myeloid-derived suppressor cells (MDSCs), thereby promoting colorectal cancer (CRC) progression. | In vitro, in vivo, mice | Treatment with M13-phage-loaded AgNPs could mop up F. nucleatum in the gut, resulting in non-amplification in MDSCs at the tumor sites. | Dong et al. [137] | |
| Shigella spp. | Shigella-specific bacteriophages: vB_SflS-ISF001, vB_SsoS-ISF002, and a cocktail of both | S. sonnei and S. flexneri causing human acute gastrointestinal infections | In vitro, cultures | More than 85% of the ESBL-positive and -negative isolates of S. sonnei and S. flexneri were inhibited by the phage cocktail (vB_SflS-ISF001 and vB_SsoS-ISF002.) | Shahin et al. [138] |
| Klebsiella pneumoniae | Lytic five-phage combination | Inflammatory bowel disease (IBD)-associated K. pneumoniae (Kp) strains | In vivo, mice | Suppression of colitis in mice | Federic et al. [139] |
| Commercial bacteriophage preparations | K. pneumoniae strains isolated from children with functional gastrointestinal disorders (FGIDs) | In vitro, spot test | Phages show negligible lytic activity, indicating the need for a more radical approach to eradicating K. pneumoniae in children with FGIDs. | Grigorova et al. [140] | |
| Listeria monocytogenes | Bacteriophage cocktail (Foodborne Outbreak Pill (FOP)) | L. monocytogenes | In vitro, simulated ilium and colon conditions | Protection against L. monocytogenes infecting the human gastrointestinal tract without causing dysbiosis. | Jakobsen et al. [142] |
| Ruminococcus gnavus | Six bacteriophages | Mucin-degrading bacterium R. gnavus from the human gut | In vivo, mice | Results show the coexistence of phages with R. gnavus in the human gut microbiome. | Buttimer et al. [144] |
| Campylobacter spp. | Double-stranded phages (Φ 16-izsam and Φ 7-izsam) | C. jejuni associated with broilers | In vivo, broilers | Phage administration showed a significant one to two log reduction in C. jejuni counts on the cecal content compared with the control group after sacrifice. The lowest colony count was, however, observed with an MOI of 0.1 of Φ 16-izsam. | D’Angelantonio et al. [146] |
| Bacteriophages φ4, φ44, φ22, φCj1, φ198, and φ287 | C. jejuni associated with broilers | In vitro, in vivo, broilers | Demonstrated the susceptibility of a significant number of the multi-resistant Campylobacter spp. to the phage isolates, which had a lytic spectrum of 6, 4, 4, 3, 8, and 7, respectively. | Nowaczek et al. [147] | |
| General | Chitosan-encapsulated bacteriophage cocktail | S. enterica, S. flexneri, and E. coli gastrointestinal infections | In vivo, rats | Reduction in positive cultures from stools of the group receiving the chitosan-encapsulated bacteriophage cocktail was observed after two days. | Rahimzade et al. [148] |
7. Roles in Preventing and Treating Specific Gut Microbiota-Related Diseases
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vemuri, R.; Shankar, E.M.; Chieppa, M.; Eri, R.; Kavanagh, K. Beyond Just Bacteria: Functional Biomes in the Gut Ecosystem Including Virome, Mycobiome, Archaeome and Helminths. Microorganisms 2020, 8, 483. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Sig. Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Young, R.; Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 135–144. [Google Scholar] [CrossRef]
- Sutton, T.D.S.; Hill, C. Gut Bacteriophage: Current Understanding and Challenges. Front. Endocrinol. 2019, 10, 784. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Popescu, M.; Van Belleghem, J.D.; Khosravi, A.; Bollyky, P.L. Bacteriophages and the Immune System. Annu. Rev. Virol. 2021, 8, 415–435. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Segal, J.P.; Carding, S.R.; Hart, A.L.; Hold, G.L. The gut virome: The ‘missing link’ between gut bacteria and host immunity? Ther. Adv. Gastroenterol. 2019, 12, 1756284819836620. [Google Scholar] [CrossRef]
- Moran-Ramos, S.; Lopez-Contreras, B.E.; Villarruel-Vazquez, R.; Ocampo-Medina, E.; Macias-Kauffer, L.; Martinez-Medina, J.N.; Villamil-Ramirez, H.; León-Mimila, P.; Del Rio-Navarro, B.E.; Ibarra-Gonzalez, I.; et al. Environmental and intrinsic factors shaping gut microbiota composition and diversity and its relation to metabolic health in children and early adolescents: A population-based study. Gut Microbes 2020, 11, 900–917. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. Gut microbiota influence in type 2 diabetes mellitus (T2DM). Gut Pathog. 2021, 13, 50. [Google Scholar] [CrossRef]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- Park, J.; Kim, C.H. Regulation of common neurological disorders by gut microbial metabolites. Exp. Mol. Med. 2021, 53, 1821–1833. [Google Scholar] [CrossRef]
- Muralitharan, R.R.; Jama, H.A.; Xie, L.; Peh, A.; Snelson, M.; Marques, F.Z. Microbial Peer Pressure: The Role of the Gut Microbiota in Hypertension and Its Complications. Hypertension 2020, 76, 1674–1687. [Google Scholar] [CrossRef]
- Brown, J.M.; Hazen, S.L. Microbial modulation of cardiovascular disease. Nat. Rev. Microbiol. 2018, 16, 171–181. [Google Scholar] [CrossRef]
- Zuo, K.; Liu, X.; Wang, P.; Jiao, J.; Han, C.; Liu, Z.; Yin, X.; Li, J.; Yang, X. Metagenomic data-mining reveals enrichment of trimethylamine-N-oxide synthesis in gut microbiome in atrial fibrillation patients. BMC Genom. 2020, 21, 526. [Google Scholar] [CrossRef]
- Breton, J.; Galmiche, M.; Déchelotte, P. Dysbiotic Gut Bacteria in Obesity: An Overview of the Metabolic Mechanisms and Therapeutic Perspectives of Next-Generation Probiotics. Microorganisms 2022, 10, 452. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Meaden, S. Understanding Bacteriophage Specificity in Natural Microbial Communities. Viruses 2013, 5, 806–823. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Nobs, S.P.; Elinav, E. Phages and their potential to modulate the microbiome and immunity. Cell Mol. Immunol. 2021, 18, 889–904. [Google Scholar] [CrossRef] [PubMed]
- AL-Ishaq, R.K.; Skariah, S.; Büsselberg, D. Bacteriophage Treatment: Critical Evaluation of Its Application on World Health Organization Priority Pathogens. Viruses 2020, 13, 51. [Google Scholar] [CrossRef] [PubMed]
- Sabino, J.; Hirten, R.P.; Colombel, J.-F. Review article: Bacteriophages in gastroenterology-from biology to clinical applications. Aliment. Pharmacol. Ther. 2020, 51, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome Composition and Its Impact on the Development of Allergic Diseases. Front. Immunol. 2020, 11, 700. [Google Scholar] [CrossRef]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.; Brugère, J.-F. Archaea and the human gut: New beginning of an old story. World J. Gastroenterol. 2014, 20, 16062–16078. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Nayfach, S.; Boland, M.; Strozzi, F.; Beracochea, M.; Shi, Z.; Pollard, K.; Sakharova, E.; Parks, D.; Philip, H.; et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021, 39, 105–114. [Google Scholar] [CrossRef]
- Pérez, J.C. Fungi of the human gut microbiota: Roles and significance. Int. J. Med. Microbiol. 2021, 311, 151490. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multi–sensitized atopy and T–cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Arévalo, A.; Stiemsma, L.; Dimitriu, P.; Chico, M.E.; Loor, S.; Vaca, M.; Boutin, R.C.T.; Morien, E.; Jin, M.; et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2018, 142, 424–434.e10. [Google Scholar] [CrossRef]
- Fiers, W.D.; Gao, I.H.; Iliev, I.D. Gut Mycobiota Under Scrutiny: Fungal Symbionts or Environmental Transients? Curr. Opin. Microbiol. 2019, 50, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Hill, C. Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host Microbe 2019, 25, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The gut virome: A new microbiome component in health and disease. eBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.R.; Janowski, A.B.; Zhao, G.; Barouch, D.; Wang, D. Hyperexpansion of RNA Bacteriophage Diversity. PLoS Biol. 2016, 14, e1002409. [Google Scholar] [CrossRef]
- Zuo, T.; Sun, Y.; Wan, Y.; Yeoh, Y.K.; Zhang, F.; Cheung, C.P.; Chen, N.; Luo, J.; Wang, W.; Sung, J.J.Y.; et al. Human-Gut-DNA Virome Variations across Geography, Ethnicity, and Urbanization. Cell Host Microbe 2020, 28, 741–751.e4. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Magwira, C.A.; Kullin, B.; Lewandowski, S.; Rodgers, A.; Reid, S.J.; Abratt, V.R. Diversity of faecal oxalate-degrading bacteria in black and white South African study groups: Insights into understanding the rarity of urolithiasis in the black group. J. Appl. Microbiol. 2012, 113, 418–428. [Google Scholar] [CrossRef]
- Brown, E.M.; Clardy, J.; Xavier, R.J. Gut microbiome lipid metabolism and its impact on host physiology. Cell Host Microbe 2023, 31, 173–186. [Google Scholar] [CrossRef]
- Shanahan, M.T.; Carroll, I.M.; Gulati, A.S. Critical design aspects involved in the study of Paneth cells and the intestinal microbiota. Gut Microbes 2014, 5, 208–214. [Google Scholar] [CrossRef]
- Fan, P.; Liu, P.; Song, P.; Chen, X.; Ma, X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep. 2017, 7, 43412. [Google Scholar] [CrossRef]
- Laparra, J.M.; Sanz, Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol. Res. 2010, 61, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Mariotti, F.; Huneau, J.F.; Tomé, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007, 33, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Shearer, M.J.; Newman, P. Recent trends in the metabolism and cell biology of vitamin K with special reference to vitamin K cycling and MK-4 biosynthesis. J. Lipid Res. 2014, 55, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Pérez, O.; Cruz-Ramón, V.; Chinchilla-López, P.; Méndez-Sánchez, N. The Role of the Gut Microbiota in Bile Acid Metabolism. Ann. Hepatol. 2017, 16, S21–S26. [Google Scholar] [CrossRef]
- Marín, L.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Bioavailability of Dietary Polyphenols and Gut Microbiota Metabolism: Antimicrobial Properties. BioMed Res. Int. 2015, 2015, 905215. [Google Scholar] [CrossRef]
- Belkaid, Y.; Harrison, O.J. Homeostatic immunity and the microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef]
- Jiao, Y.; Wu, L.; Huntington, N.D.; Zhang, X. Crosstalk Between Gut Microbiota and Innate Immunity and Its Implication in Autoimmune Diseases. Front. Immunol. 2020, 11, 282. [Google Scholar] [CrossRef]
- Zhao, Q.; Elson, C.O. Adaptive immune education by gut microbiota antigens. Immunology 2018, 154, 28–37. [Google Scholar] [CrossRef]
- Martin-Gallausiaux, C.; Béguet-Crespel, F.; Marinelli, L.; Jamet, A.; Ledue, F.; Blottière, H.M.; Lapaque, N. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci. Rep. 2018, 8, 9742. [Google Scholar] [CrossRef]
- den Besten, G.; Gerding, A.; van Dijk, T.H.; Ciapaite, J.; Bleeker, A.; van Eunen, K.; Havinga, R.; Groen, A.K.; Reijngoud, D.-J.; Bakker, B.M. Protection against the Metabolic Syndrome by Guar Gum-Derived Short-Chain Fatty Acids Depends on Peroxisome Proliferator-Activated Receptor γ and Glucagon-Like Peptide-1. PLoS ONE 2015, 10, e0136364. [Google Scholar] [CrossRef]
- Oviedo-Boyso, J.; Bravo-Patiño, A.; Baizabal-Aguirre, V.M. Collaborative Action of Toll-Like and Nod-Like Receptors as Modulators of the Inflammatory Response to Pathogenic Bacteria. Mediat. Inflamm. 2014, 2014, 432785. [Google Scholar] [CrossRef] [PubMed]
- Bunker, J.J.; Flynn, T.M.; Koval, J.C.; Shaw, D.G.; Meisel, M.; McDonald, B.D.; Ishizuka, I.E.; Dent, A.L.; Wilson, P.C.; Jabri, B.; et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015, 43, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.A.; Ryu, J.H.; Jo, Y.; Hong, C. The role of gut microbiota in T cell immunity and immune mediated disorders. Int. J. Biol. Sci. 2023, 19, 1178–1191. [Google Scholar] [CrossRef]
- Li, Y.; Ye, Z.; Zhu, J.; Fang, S.; Meng, L.; Zhou, C. Effects of Gut Microbiota on Host Adaptive Immunity Under Immune Homeostasis and Tumor Pathology State. Front. Immunol. 2022, 13, 844335. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.; Cham, C.; et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Fu, J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Ubeda, C.; Djukovic, A.; Isaac, S. Roles of the intestinal microbiota in pathogen protection. Clin. Trans. Immunol. 2017, 6, e128. [Google Scholar] [CrossRef]
- Qadir, A.; Hashmi, M.Z.; Mahmood, A. Xenobiotics, Types, and Mode of Action. In Xenobiotics in the Soil Environment: Monitoring, Toxicity and Management; Springer: Cham, Switzerland, 2017; Volume 49, pp. 1–7. ISBN 978-3-319-47743-5. [Google Scholar]
- Collins, S.L.; Patterson, A.D. The gut microbiome: An orchestrator of xenobiotic metabolism. Acta Pharm. Sin. B 2020, 10, 19–32. [Google Scholar] [CrossRef]
- Pant, A.; Maiti, T.K.; Mahajan, D.; Das, B. Human Gut Microbiota and Drug Metabolism. Microb. Ecol. 2022, 86, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [PubMed]
- ElRakaiby, M.; Dutilh, B.E.; Rizkallah, M.R.; Boleij, A.; Cole, J.N.; Aziz, R.K. Pharmacomicrobiomics: The Impact of Human Microbiome Variations on Systems Pharmacology and Personalized Therapeutics. OMICS 2014, 18, 402–414. [Google Scholar] [CrossRef]
- Aziz, R.K. Rethinking Pharmacogenomics in an Ecosystem: Drug-microbiome Interactions, Pharmacomicrobiomics, and Personalized Medicine for the Human Supraorganism. Curr. Pharmacogenom. Pers. Med. 2012, 10, 258–261. [Google Scholar] [CrossRef]
- Yoo, D.-H.; Kim, I.S.; Le, T.K.V.; Jung, I.-H.; Yoo, H.H.; Kim, D.-H. Gut Microbiota-Mediated Drug Interactions between Lovastatin and Antibiotics. Drug Metab. Dispos. 2014, 42, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Dhurjad, P.; Dhavaliker, C.; Gupta, K.; Sonti, R. Exploring Drug Metabolism by the Gut Microbiota: Modes of Metabolism and Experimental Approaches. Drug Metab. Dispos. 2022, 50, 224–234. [Google Scholar] [CrossRef]
- Chaudhari, S.N.; Luo, J.N.; Harris, D.A.; Aliakbarian, H.; Yao, L.; Paik, D.; Subramaniam, R.; Adhikari, A.A.; Vernon, A.H.; Kiliç, A.; et al. A microbial metabolite remodels the gut-liver axis following bariatric surgery. Cell Host Microbe 2021, 29, 408–424.e7. [Google Scholar] [CrossRef]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.-A.; Mani, S.; et al. Alleviating Cancer Drug Toxicity by Inhibiting a Bacterial Enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
- Gieryńska, M.; Szulc-Dąbrowska, L.; Struzik, J.; Mielcarska, M.B.; Gregorczyk-Zboroch, K.P. Integrity of the Intestinal Barrier: The Involvement of Epithelial Cells and Microbiota—A Mutual Relationship. Animals 2022, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Han, J.; Li, L.; Wang, Y.; Li, Y.; Zhang, S. Claudin Family Participates in the Pathogenesis of Inflammatory Bowel Diseases and Colitis-Associated Colorectal Cancer. Front. Immunol. 2019, 10, 1441. [Google Scholar] [CrossRef] [PubMed]
- Okumura, R.; Takeda, K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Tailed Bacteriophages: The Order Caudovirales. Adv. Virus Res. 1998, 51, 135–201. [Google Scholar] [CrossRef]
- Marvin, D.A.; Symmons, M.F.; Straus, S.K. Structure and assembly of filamentous bacteriophages. Prog. Biophys. Mol. Biol. 2014, 114, 80–122. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Family—Tectiviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 317–321. ISBN 978-0-12-384684-6. [Google Scholar]
- Knezevic, P.; Adriaenssens, E.M. ICTV Virus Taxonomy Profile: Inoviridae. J. Gen. Virol. 2021, 102, 001614. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Family—Leviviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 1035–1043. ISBN 978-0-12-384684-6. [Google Scholar]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Family—Microviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 385–393. ISBN 978-0-12-384684-6. [Google Scholar]
- Pietilä, M.K.; Roine, E.; Sencilo, A.; Bamford, D.H.; Oksanen, H.M. Pleolipoviridae, a newly proposed family comprising archaeal pleomorphic viruses with single-stranded or double-stranded DNA genomes. Arch. Virol. 2016, 161, 249–256. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their Genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef]
- Callanan, J.; Stockdale, S.R.; Shkoporov, A.; Draper, L.A.; Ross, R.P.; Hill, C. Expansion of known ssRNA phage genomes: From tens to over a thousand. Sci. Adv. 2020, 6, eaay5981. [Google Scholar] [CrossRef]
- Yang, Y.; Lu, S.; Shen, W.; Zhao, X.; Shen, M.; Tan, Y.; Li, G.; Li, M.; Wang, J.; Hu, F.; et al. Characterization of the first double-stranded RNA bacteriophage infecting Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 38795. [Google Scholar] [CrossRef]
- Ilina, T.V.; Brosenitsch, T.; Sluis-Cremer, N.; Ishima, R. Retroviral RNase H: Structure, mechanism, and inhibition. Enzymes 2021, 50, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Wawrzyniak, P.; Płucienniczak, G.; Bartosik, D. The Different Faces of Rolling-Circle Replication and Its Multifunctional Initiator Proteins. Front. Microbiol. 2017, 8, 2353. [Google Scholar] [CrossRef] [PubMed]
- Salmond, G.P.C.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shang, J.; Peng, C.; Sun, Y. Phage family classification under Caudoviricetes: A review of current tools using the latest ICTV classification framework. Front. Microbiol. 2022, 13, 1032186. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, T.; Yu, M.; Chen, Y.-L.; Jin, M. The Life Cycle Transitions of Temperate Phages: Regulating Factors and Potential Ecological Implications. Viruses 2022, 14, 1904. [Google Scholar] [CrossRef]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophage Adsorption: Likelihood of Virion Encounter with Bacteria and Other Factors Affecting Rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef]
- Criel, B.; Taelman, S.; Van Criekinge, W.; Stock, M.; Briers, Y. PhaLP: A Database for the Study of Phage Lytic Proteins and Their Evolution. Viruses 2021, 13, 1240. [Google Scholar] [CrossRef]
- Oliveira, H.; São-José, C.; Azeredo, J. Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for in vivo Therapy. Viruses 2018, 10, 292. [Google Scholar] [CrossRef]
- Attama, A.; Agbo, I.; Eke, I.; Onuigbo, E.; Ogbonna, J. Bacteriophage: Clinical Applications. In Antimicrobial Research: Novel Bioknowledge and Educational Programs; Mendez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2017; ISBN 978-84-947512-0-2. [Google Scholar]
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 2019, 25, 803–814.e5. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.R.; Davis, N.; Hoyles, L. Review article: The human intestinal virome in health and disease. Aliment. Pharmacol. Ther. 2017, 46, 800–815. [Google Scholar] [CrossRef]
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; McCartney, A.L.; Neve, H.; Gibson, G.R.; Sanderson, J.D.; Heller, K.J.; van Sinderen, D. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 2014, 165, 803–812. [Google Scholar] [CrossRef]
- Martinez-Castillo, A.; Quirós, P.; Navarro, F.; Miró, E.; Muniesa, M. Shiga Toxin 2-Encoding Bacteriophages in Human Fecal Samples from Healthy Individuals. Appl. Environ. Microbiol. 2013, 79, 4862–4868. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Hewson, I.; Felts, B.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Metagenomic Analyses of an Uncultured Viral Community from Human Feces. J. Bacteriol. 2003, 185, 6220–6223. [Google Scholar] [CrossRef]
- Reyes, A.; Semenkovich, N.P.; Whiteson, K.; Rohwer, F.; Gordon, J.I. Going viral: Next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 2012, 10, 607–617. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef]
- Minot, S.; Bryson, A.; Chehoud, C.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA 2013, 110, 12450–12455. [Google Scholar] [CrossRef]
- Liang, G.; Zhao, C.; Zhang, H.; Mattei, L.; Sherrill-Mix, S.; Bittinger, K.; Kessler, L.R.; Wu, G.D.; Baldassano, R.N.; DeRusso, P.; et al. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 2020, 581, 470–474. [Google Scholar] [CrossRef]
- Kåhrström, C.T.; Pariente, N.; Weiss, U. Intestinal microbiota in health and disease. Nature 2016, 535, 47. [Google Scholar] [CrossRef]
- Olovo, C.V.; Huang, X.; Zheng, X.; Xu, M. Faecal microbial biomarkers in early diagnosis of colorectal cancer. J. Cell. Mol. Med. 2021, 25, 10783–10797. [Google Scholar] [CrossRef]
- Shuwen, H.; Kefeng, D. Intestinal phages interact with bacteria and are involved in human diseases. Gut Microbes 2022, 14, 2113717. [Google Scholar] [CrossRef]
- Nikolich, M.P.; Filippov, A.A. Bacteriophage Therapy: Developments and Directions. Antibiotics 2020, 9, 135. [Google Scholar] [CrossRef]
- Jaiswal, A.; Koley, H.; Ghosh, A.; Palit, A.; Sarkar, B. Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes Infect. 2013, 15, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.; Koley, H.; Mitra, S.; Saha, D.R.; Sarkar, B. Comparative analysis of different oral approaches to treat Vibrio cholerae infection in adult mice. Int. J. Med. Microbiol. 2014, 304, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.W.; Shin, T.H.; Kim, J.H.; Shin, S.P.; Han, J.E.; Heo, G.J.; De Zoysa, M.; Shin, G.W.; Chai, J.Y.; Park, S.C. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J. Infect. Dis. 2014, 210, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Yen, M.; Cairns, L.S.; Camilli, A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 2017, 8, 14187. [Google Scholar] [CrossRef] [PubMed]
- Nasr-Eldin, M.A.; EL-Maaty, S.A.A.; EL-Dougdoug, K.A.; Hazaa, M.M.; Abdel-mageed, A.H. Characterization and development of a phage cocktail for Escherichia coli causing gastrointestinal diseases. J. Basic Environ. Sci. 2018, 5, 115–122. [Google Scholar]
- Abdulamir, A.S.; Jassim, S.A.; Abu Bakar, F. Novel approach of using a cocktail of designed bacteriophages against gut pathogenic E. colifor bacterial load biocontrol. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.A.; Abo Kamer, A.M.; Nosair, A.M.; Al-Madboly, L.A. Exploring the potential efficacy of phage therapy for biocontrol of foodborne pathogenic extensively drug-resistant Escherichia coli in gastrointestinal tract of rat model. Life Sci. 2023, 315, 121362. [Google Scholar] [CrossRef]
- Bourdin, G.; Navarro, A.; Sarker, S.A.; Pittet, A.-C.; Qadri, F.; Sultana, S.; Cravioto, A.; Talukder, K.A.; Reuteler, G.; Brüssow, H. Coverage of diarrhoea-associated Escherichia coli isolates from different origins with two types of phage cocktails. Microb. Biotechnol. 2014, 7, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Aleshkin, A.V.; Rubalskii, E.O.; Volozhantsev, N.V.; Verevkin, V.V.; Svetoch, E.A.; Kiseleva, I.A.; Bochkareva, S.S.; Borisova, O.Y.; Popova, A.V.; Bogun, A.G.; et al. A small-scale experiment of using phage-based probiotic dietary supplement for prevention of E. coli traveler’s diarrhea. Bacteriophage 2015, 5, e1074329. [Google Scholar] [CrossRef] [PubMed]
- Vahedi, A.; Soltan Dallal, M.M.; Douraghi, M.; Nikkhahi, F.; Rajabi, Z.; Yousefi, M.; Mousavi, M. Isolation and identification of specific bacteriophage against enteropathogenic Escherichia coli (EPEC) and in vitro and in vivo characterization of bacteriophage. FEMS Microbiol. Lett. 2018, 365, fny136. [Google Scholar] [CrossRef]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T.; et al. Oral Phage Therapy of Acute Bacterial Diarrhea with Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Galtier, M.; Sordi, L.D.; Sivignon, A.; de Vallée, A.; Maura, D.; Neut, C.; Rahmouni, O.; Wannerberger, K.; Darfeuille-Michaud, A.; Desreumaux, P.; et al. Bacteriophages Targeting Adherent Invasive Escherichia coli Strains as a Promising New Treatment for Crohn’s Disease. J. Crohn’s Colitis 2017, 11, 840–847. [Google Scholar] [CrossRef]
- Cieplak, T.; Soffer, N.; Sulakvelidze, A.; Nielsen, D.S. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 2018, 9, 391–399. [Google Scholar] [CrossRef]
- Gindin, M.; Febvre, H.P.; Rao, S.; Wallace, T.C.; Weir, T.L. Bacteriophage for Gastrointestinal Health (PHAGE) Study: Evaluating the Safety and Tolerability of Supplemental Bacteriophage Consumption. J. Am. Coll. Nutr. 2019, 38, 68–75. [Google Scholar] [CrossRef]
- Grubb, D.S.; Wrigley, S.D.; Freedman, K.E.; Wei, Y.; Vazquez, A.R.; Trotter, R.E.; Wallace, T.C.; Johnson, S.A.; Weir, T.L. PHAGE-2 Study: Supplemental Bacteriophages Extend Bifidobacterium animalis subsp. lactis BL04 Benefits on Gut Health and Microbiota in Healthy Adults. Nutrients 2020, 12, 2474. [Google Scholar] [CrossRef]
- Alomari, M.M.M.; Dec, M.; Nowaczek, A.; Puchalski, A.; Wernicki, A.; Kowalski, C.; Urban-Chmiel, R. Therapeutic and Prophylactic Effect of the Experimental Bacteriophage Treatment to Control Diarrhea Caused by E. coli in Newborn Calves. ACS Infect. Dis. 2021, 7, 2093–2101. [Google Scholar] [CrossRef]
- Hsu, B.B.; Way, J.C.; Silver, P.A. Stable Neutralization of a Virulence Factor in Bacteria Using Temperate Phage in the Mammalian Gut. mSystems 2020, 5, e00013-20. [Google Scholar] [CrossRef]
- Cepko, L.C.S.; Garling, E.E.; Dinsdale, M.J.; Scott, W.P.; Bandy, L.; Nice, T.; Faber-Hammond, J.; Mellies, J.L. Myoviridae phage PDX kills enteroaggregative Escherichia coli without human microbiome dysbiosis. J. Med. Microbiol. 2020, 69, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Green, S.I.; Gu Liu, C.; Yu, X.; Gibson, S.; Salmen, W.; Rajan, A.; Carter, H.E.; Clark, J.R.; Song, X.; Ramig, R.F.; et al. Targeting of Mammalian Glycans Enhances Phage Predation in the Gastrointestinal Tract. mBio 2021, 12, e03474-20. [Google Scholar] [CrossRef]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepiński, P.; Douce, G.R.; Clokie, M.R.J. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth in vitro and Proliferation in vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Selle, K.; Fletcher, J.R.; Tuson, H.; Schmitt, D.S.; McMillan, L.; Vridhambal, G.S.; Rivera, A.J.; Montgomery, S.A.; Fortier, L.-C.; Barrangou, R.; et al. In vivo Targeting of Clostridioides difficile Using Phage-Delivered CRISPR-Cas3 Antimicrobials. mBio 2020, 11, e00019-20. [Google Scholar] [CrossRef] [PubMed]
- Dallal, M.M.S.; Nikkhahi, F.; Alimohammadi, M.; Douraghi, M.; Rajabi, Z.; Foroushani, A.R.; Azimi, A.; Fardsanei, F. Phage Therapy as an Approach to Control Salmonella enterica serotype Enteritidis Infection in Mice. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190290. [Google Scholar] [CrossRef]
- Thanki, A.M.; Hooton, S.; Whenham, N.; Salter, M.G.; Bedford, M.R.; O’Neill, H.V.M.; Clokie, M.R.J. A bacteriophage cocktail delivered in feed significantly reduced Salmonella colonization in challenged broiler chickens. Emerg. Microbes Infect. 2023, 12, 2217947. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Abbeele, P.V.D.; Duysburgh, C.; Verstrepen, L.; Das, C.R.; Marzorati, M.; Sulakvelidze, A. A Bacteriophage Cocktail Eliminates Salmonella Typhimurium from the Human Colonic Microbiome while Preserving Cytokine Signaling and Preventing Attachment to and Invasion of Human Cells by Salmonella in vitro. J. Food Prot. 2019, 82, 1336–1349. [Google Scholar] [CrossRef]
- Nale, J.Y.; Vinner, G.K.; Lopez, V.C.; Thanki, A.M.; Phothaworn, P.; Thiennimitr, P.; Garcia, A.; AbuOun, M.; Anjum, M.F.; Korbsrisate, S.; et al. An Optimized Bacteriophage Cocktail Can Effectively Control Salmonella in vitro and in Galleria mellonella. Front. Microbiol. 2021, 11, 609955. [Google Scholar] [CrossRef]
- Pelyuntha, W.; Yafa, A.; Ngasaman, R.; Yingkajorn, M.; Chukiatsiri, K.; Champoochana, N.; Vongkamjan, K. Oral Administration of a Phage Cocktail to Reduce Salmonella Colonization in Broiler Gastrointestinal Tract—A Pilot Study. Animals 2022, 12, 3087. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.-W.; Dong, X.; Pan, P.; Chen, K.-W.; Fan, J.-X.; Cheng, S.-X.; Zhang, X.-Z. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019, 3, 717–728. [Google Scholar] [CrossRef]
- Dong, X.; Pan, P.; Zheng, D.-W.; Bao, P.; Zeng, X.; Zhang, X.-Z. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 2020, 6, eaba1590. [Google Scholar] [CrossRef]
- Shahin, K.; Bouzari, M.; Komijani, M.; Wang, R. A New Phage Cocktail Against Multidrug, ESBL-Producer Isolates of Shigella sonnei and Shigella flexneri with Highly Efficient Bacteriolytic Activity. Microb. Drug Resist. 2020, 26, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, A.; et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022, 185, 2879–2898.e24. [Google Scholar] [CrossRef]
- Grigorova, E.V.; Rychkova, L.V.; Belkova, N.L.; Nemchenko, U.M.; Savelkaeva, M.V.; Kungurtseva, E.A.; Voropaeva, N.M. Evaluation of the sensitivity of bacteriophage preparations to Klebsiella pneumoniae strains isolated from the colon microbiota in children with functional gastrointestinal disorders. Klin. Lab. Diagn. 2021, 66, 217–222. [Google Scholar] [CrossRef]
- De Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, R.R.; Trinh, J.T.; Bomholtz, L.; Brok-Lauridsen, S.K.; Sulakvelidze, A.; Nielsen, D.S. A Bacteriophage Cocktail Significantly Reduces Listeria monocytogenes without Deleterious Impact on the Commensal Gut Microbiota under Simulated Gastrointestinal Conditions. Viruses 2022, 14, 190. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Littmann, E.R.; Fontana, E.; Moody, T.U.; Kohout, C.E.; Gjonbalaj, M.; Eaton, V.; Seok, R.; Leiner, I.M.; Pamer, E.G. Functional and Genomic Variation between Human-Derived Isolates of Lachnospiraceae Reveals Inter- and Intra-Species Diversity. Cell Host Microbe 2020, 28, 134–146.e4. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Khokhlova, E.V.; Stein, L.; Hueston, C.M.; Govi, B.; Draper, L.A.; Ross, R.P.; Shkoporov, A.N.; Hill, C. Temperate bacteriophages infecting the mucin-degrading bacterium Ruminococcus gnavus from the human gut. Gut Microbes 2023, 15, 2194794. [Google Scholar] [CrossRef]
- Olson, E.G.; Micciche, A.C.; Rothrock, M.J.; Yang, Y.; Ricke, S.C. Application of Bacteriophages to Limit Campylobacter in Poultry Production. Front. Microbiol. 2022, 12, 458721. [Google Scholar] [CrossRef] [PubMed]
- D’Angelantonio, D.; Scattolini, S.; Boni, A.; Neri, D.; Di Serafino, G.; Connerton, P.; Connerton, I.; Pomilio, F.; Di Giannatale, E.; Migliorati, G.; et al. Bacteriophage Therapy to Reduce Colonization of Campylobacter jejuni in Broiler Chickens before Slaughter. Viruses 2021, 13, 1428. [Google Scholar] [CrossRef] [PubMed]
- Nowaczek, A.; Urban-Chmiel, R.; Dec, M.; Puchalski, A.; Stępień-Pyśniak, D.; Marek, A.; Pyzik, E. Campylobacter spp. and bacteriophages from broiler chickens: Characterization of antibiotic susceptibility profiles and lytic bacteriophages. Microbiologyopen 2019, 8, e00784. [Google Scholar] [CrossRef] [PubMed]
- Rahimzadeh, G.; Saeedi, M.; Moosazadeh, M.; Hashemi, S.M.H.; Babaei, A.; Rezai, M.S.; Kamel, K.; Asare-Addo, K.; Nokhodchi, A. Encapsulation of bacteriophage cocktail into chitosan for the treatment of bacterial diarrhea. Sci. Rep. 2021, 11, 15603. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological applications of bacteriophages: State of the art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Shlezinger, M.; Khalifa, L.; Houri-Haddad, Y.; Coppenhagen-Glazer, S.; Resch, G.; Que, Y.-A.; Beyth, S.; Dorfman, E.; Hazan, R.; Beyth, N. Phage Therapy: A New Horizon in the Antibacterial Treatment of Oral Pathogens. Curr. Top. Med. Chem. 2017, 17, 1199–1211. [Google Scholar] [CrossRef]
- Baghi, H.B.; Naghili, B.; Shanehbandi, D.; Leylabadlo, H.E. Evaluation of a human gut-associated phage and gut dominant microbial phyla in the metabolic syndrome. Clin. Nutr. ESPEN 2022, 50, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Międzybrodzki, R.; Borysowski, J.; Dąbrowska, K.; Wierzbicki, P.; Ohams, M.; Korczak-Kowalska, G.; Olszowska-Zaremba, N.; Łusiak-Szelachowska, M.; Kłak, M.; et al. Phage as a modulator of immune responses: Practical implications for phage therapy. Adv. Virus Res. 2012, 83, 41–71. [Google Scholar] [CrossRef]
- Foglizzo, V.; Marchiò, S. Bacteriophages as Therapeutic and Diagnostic Vehicles in Cancer. Pharmaceuticals 2021, 14, 161. [Google Scholar] [CrossRef]
- Kabwe, M.; Meehan-Andrews, T.; Ku, H.; Petrovski, S.; Batinovic, S.; Chan, H.T.; Tucci, J. Lytic Bacteriophage EFA1 Modulates HCT116 Colon Cancer Cell Growth and Upregulates ROS Production in an Enterococcus faecalis Co-culture System. Front. Microbiol. 2021, 12, 650849. [Google Scholar] [CrossRef]
- Kabwe, M.; Dashper, S.; Tucci, J. The Microbiome in Pancreatic Cancer-Implications for Diagnosis and Precision Bacteriophage Therapy for This Low Survival Disease. Front. Cell Infect. Microbiol. 2022, 12, 871293. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, K.; Duan, H.; Du, Y.; Ye, J. Phage-based peptides for pancreatic cancer diagnosis and treatment: Alternative approach. Front. Microbiol. 2023, 14, 1231503. [Google Scholar] [CrossRef]
- Hong, D.; Zhang, C.; Wu, W.; Lu, X.; Zhang, L. Modulation of the gut–brain axis via the gut microbiota: A new era in treatment of amyotrophic lateral sclerosis. Front. Neurol. 2023, 14, 1133546. [Google Scholar] [CrossRef] [PubMed]
- Ghadge, G.D.; Kay, B.K.; Drigotas, C.; Roos, R.P. Single chain variable fragment antibodies directed against SOD1 ameliorate disease in mutant SOD1 transgenic mice. Neurobiol. Dis. 2019, 121, 131–137. [Google Scholar] [CrossRef] [PubMed]


| Morphology | Genomic Properties | Life Cycle |
|---|---|---|
Caudovirales
| DNA Phages
| Temperate |
| Filamentous | RNA Phages
| Lytic |
| Tectiviridae | Retroviruses | Lysogenic |
| Inoviridae | Circular Replicating Phages | |
| Leviviridae | Temperate Phages | |
| Microviridae | Virulent Phages | |
| Pleolipoviridae |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emencheta, S.C.; Olovo, C.V.; Eze, O.C.; Kalu, C.F.; Berebon, D.P.; Onuigbo, E.B.; Vila, M.M.D.C.; Balcão, V.M.; Attama, A.A. The Role of Bacteriophages in the Gut Microbiota: Implications for Human Health. Pharmaceutics 2023, 15, 2416. https://doi.org/10.3390/pharmaceutics15102416
Emencheta SC, Olovo CV, Eze OC, Kalu CF, Berebon DP, Onuigbo EB, Vila MMDC, Balcão VM, Attama AA. The Role of Bacteriophages in the Gut Microbiota: Implications for Human Health. Pharmaceutics. 2023; 15(10):2416. https://doi.org/10.3390/pharmaceutics15102416
Chicago/Turabian StyleEmencheta, Stephen C., Chinasa V. Olovo, Osita C. Eze, Chisom F. Kalu, Dinebari P. Berebon, Ebele B. Onuigbo, Marta M. D. C. Vila, Victor M. Balcão, and Anthony A. Attama. 2023. "The Role of Bacteriophages in the Gut Microbiota: Implications for Human Health" Pharmaceutics 15, no. 10: 2416. https://doi.org/10.3390/pharmaceutics15102416







