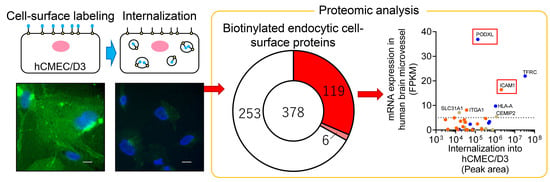
Identification of Cell-Surface Proteins Endocytosed by Human Brain Microvascular Endothelial Cells In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Internalization Assay
2.3. Fluorescence Microscopy
2.4. Silver Staining
2.5. Quantitative Proteome Analysis
2.6. Identification of Internalized Proteins
2.7. Immunohistochemistry
2.8. Internalization of Antibody-Labeled Cell-Surface Protein in the Cells
2.9. Statistical Analysis
3. Results
3.1. Detection of Biotinylated Proteins in hCMEC/D3 Cells and HUVECs
3.2. Identification of “Biotinylated Endocytic Cell-Surface Proteins” in hCMEC/D3 Cells and HUVECs
3.3. Identification of Selected Endocytic Cell-Surface Proteins in hCMEC/D3 Cells and HUVECs
3.4. Identification of Brain Microvessel Endothelial Cell-Selective Endocytic Cell-Surface Proteins
3.5. Internalization of PODXL into hCMEC/D3 Cells
3.6. Immunohistochemical Analysis of PODXL in Human Cerebral Cortex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fishman, J.B.; Rubin, J.B.; Handrahan, J.V.; Connor, J.R.; Fine, R.E. Receptor-mediated transcytosis of transferrin across the blood-brain barrier. J. Neurosci. Res. 1987, 18, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Friden, P.M.; Walus, L.R.; Musso, G.F.; Taylor, M.A.; Malfroy, B.; Starzyk, R.M. Anti-transferrin receptor antibody and antibody-drug conjugates cross the blood-brain barrier. Proc. Natl. Acad. Sci. USA 1991, 88, 4771–4775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuyama, T.; Eto, Y.; Sakai, N.; Minami, K.; Yamamoto, T.; Sonoda, H.; Yamaoka, M.; Tachibana, K.; Hirato, T.; Sato, Y. Iduronate-2-Sulfatase with Anti-human Transferrin Receptor Antibody for Neuropathic Mucopolysaccharidosis II: A Phase 1/2 Trial. Mol. Ther. 2019, 27, 456–464. [Google Scholar] [CrossRef] [Green Version]
- Sonoda, H.; Morimoto, H.; Yoden, E.; Koshimura, Y.; Kinoshita, M.; Golovina, G.; Takagi, H.; Yamamoto, R.; Minami, K.; Mizoguchi, A.; et al. A Blood-Brain-Barrier-Penetrating Anti-human Transferrin Receptor Antibody Fusion Protein for Neuronopathic Mucopolysaccharidosis II. Mol. Ther. 2018, 26, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Couch, J.A.; Yu, Y.J.; Zhang, Y.; Tarrant, J.M.; Fuji, R.N.; Meilandt, W.J.; Solanoy, H.; Tong, R.K.; Hoyte, K.; Luk, W.; et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood-brain barrier. Sci. Transl. Med. 2013, 5, 183ra57. [Google Scholar] [CrossRef]
- Lee, H.J.; Engelhardt, B.; Lesley, J.; Bickel, U.; Pardridge, W.M. Targeting rat anti-mouse transferrin receptor monoclonal antibodies through blood-brain barrier in mouse. J. Pharmacol. Exp. Ther. 2000, 292, 1048–1052. [Google Scholar]
- Demeule, M.; Currie, J.C.; Bertrand, Y.; Che, C.; Nguyen, T.; Regina, A.; Gabathuler, R.; Castaigne, J.P.; Beliveau, R. Involvement of the low-density lipoprotein receptor-related protein in the transcytosis of the brain delivery vector angiopep-2. J. Neurochem. 2008, 106, 1534–1544. [Google Scholar] [CrossRef]
- Pardridge, W.M.; Boado, R.J. Reengineering biopharmaceuticals for targeted delivery across the blood-brain barrier. Methods Enzymol. 2012, 503, 269–292. [Google Scholar]
- Zuchero, Y.J.; Chen, X.; Bien-Ly, N.; Bumbaca, D.; Tong, R.K.; Gao, X.; Zhang, S.; Hoyte, K.; Luk, W.; Huntley, M.A.; et al. Discovery of Novel Blood-Brain Barrier Targets to Enhance Brain Uptake of Therapeutic Antibodies. Neuron 2016, 89, 70–82. [Google Scholar] [CrossRef] [Green Version]
- Anraku, Y.; Kuwahara, H.; Fukusato, Y.; Mizoguchi, A.; Ishii, T.; Nitta, K.; Matsumoto, Y.; Toh, K.; Miyata, K.; Uchida, S.; et al. Glycaemic control boosts glucosylated nanocarrier crossing the BBB into the brain. Nat. Commun. 2017, 8, 1001. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, H.; Tachikawa, M.; Yagi, Y.; Umetsu, M.; Nurdin, A.; Miyauchi, E.; Watanabe, M.; Uchida, Y.; Terasaki, T. Cluster of Differentiation 46 Is the Major Receptor in Human Blood-Brain Barrier Endothelial Cells for Uptake of Exosomes Derived from Brain-Metastatic Melanoma Cells (SK-Mel-28). Mol. Pharm. 2019, 16, 292–304. [Google Scholar] [CrossRef]
- Bitsikas, V.; Correa, I.R., Jr.; Nichols, B.J. Clathrin-independent pathways do not contribute significantly to endocytic flux. Elife 2014, 3, e03970. [Google Scholar] [CrossRef]
- Hormann, K.; Stukalov, A.; Muller, A.C.; Heinz, L.X.; Superti-Furga, G.; Colinge, J.; Bennett, K.L. A Surface Biotinylation Strategy for Reproducible Plasma Membrane Protein Purification and Tracking of Genetic and Drug-Induced Alterations. J. Proteome Res. 2016, 15, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Ohtsuki, S.; Murata, S.; Katsukura, Y.; Suzuki, H.; Funaki, M.; Tachikawa, M.; Terasaki, T. Involvement of insulin-degrading enzyme in insulin- and atrial natriuretic peptide-sensitive internalization of amyloid-beta peptide in mouse brain capillary endothelial cells. J. Alzheimers Dis. 2014, 38, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Markoutsa, S.; Bahr, U.; Papasotiriou, D.G.; Hafner, A.K.; Karas, M.; Sorg, B.L. Sulfo-NHS-SS-biotin derivatization: A versatile tool for MALDI mass analysis of PTMs in lysine-rich proteins. Proteomics 2014, 14, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Hirayama-Kurogi, M.; Ito, S.; Kuno, T.; Yoneyama, T.; Obuchi, W.; Terasaki, T.; Ohtsuki, S. Large-scale multiplex absolute protein quantification of drug-metabolizing enzymes and transporters in human intestine, liver, and kidney microsomes by SWATH-MS: Comparison with MRM/SRM and HR-MRM/PRM. Proteomics 2016, 16, 2106–2117. [Google Scholar] [CrossRef]
- Masuda, T.; Hoshiyama, T.; Uemura, T.; Hirayama-Kurogi, M.; Ogata, S.; Furukawa, A.; Couraud, P.O.; Furihata, T.; Ito, S.; Ohtsuki, S. Large-Scale Quantitative Comparison of Plasma Transmembrane Proteins between Two Human Blood-Brain Barrier Model Cell Lines, hCMEC/D3 and HBMEC/cibeta. Mol. Pharm. 2019, 16, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuki, S.; Ikeda, C.; Uchida, Y.; Sakamoto, Y.; Miller, F.; Glacial, F.; Decleves, X.; Scherrmann, J.M.; Couraud, P.O.; Kubo, Y.; et al. Quantitative targeted absolute proteomic analysis of transporters, receptors and junction proteins for validation of human cerebral microvascular endothelial cell line hCMEC/D3 as a human blood-brain barrier model. Mol. Pharm. 2013, 10, 289–296. [Google Scholar] [CrossRef]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.; Li, G.; et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weksler, B.B.; Subileau, E.A.; Perriere, N.; Charneau, P.; Holloway, K.; Leveque, M.; Tricoire-Leignel, H.; Nicotra, A.; Bourdoulous, S.; Turowski, P.; et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005, 19, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Tomita, M.; Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008, 7, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Fujino, Y.; Ogata, S.; Hirayama-Kurogi, M.; Ohtsuki, S. Involvement of an Orphan Transporter, SLC22A18, in Cell Growth and Drug Resistance of Human Breast Cancer MCF7 Cells. J. Pharm. Sci. 2018, 107, 3163–3170. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Salani, B.; Passalacqua, M.; Maffioli, S.; Briatore, L.; Hamoudane, M.; Contini, P.; Cordera, R.; Maggi, D. IGF-IR internalizes with Caveolin-1 and PTRF/Cavin in HaCat cells. PLoS ONE 2010, 5, e14157. [Google Scholar] [CrossRef] [Green Version]
- Muro, S.; Wiewrodt, R.; Thomas, A.; Koniaris, L.; Albelda, S.M.; Muzykantov, V.R.; Koval, M. A novel endocytic pathway induced by clustering endothelial ICAM-1 or PECAM-1. J. Cell Sci. 2003, 116, 1599–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Noss, E.H.; Hsu, V.W.; Brenner, M.B. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J. Cell Biol. 2011, 193, 61–70. [Google Scholar] [CrossRef]
- Yawata, T.; Higashi, Y.; Kawanishi, Y.; Nakajo, T.; Fukui, N.; Fukuda, H.; Ueba, T. CD146 is highly expressed in glioma stem cells and acts as a cell cycle regulator. J. Neurooncol. 2019, 144, 21–32. [Google Scholar] [CrossRef]
- Camby, I.; Belot, N.; Lefranc, F.; Sadeghi, N.; de Launoit, Y.; Kaltner, H.; Musette, S.; Darro, F.; Danguy, A.; Salmon, I.; et al. Galectin-1 modulates human glioblastoma cell migration into the brain through modifications to the actin cytoskeleton and levels of expression of small GTPases. J. Neuropathol. Exp. Neurol. 2002, 61, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, K.; Mishima, K.; Nagashima, Y.; Asai, A.; Sanai, Y.; Kirino, T. Expression of galectin-1 mRNA correlates with the malignant potential of human gliomas and expression of antisense galectin-1 inhibits the growth of 9 glioma cells. J. Neurosci. Res. 2000, 59, 722–730. [Google Scholar] [CrossRef]
- Weksler, B.; Romero, I.A.; Couraud, P.O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS 2013, 10, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossler, K.; Neuchrist, C.; Kitz, K.; Scheiner, O.; Kraft, D.; Lassmann, H. Expression of leucocyte adhesion molecules at the human blood-brain barrier (BBB). J. Neurosci. Res. 1992, 31, 365–374. [Google Scholar] [CrossRef]
- Lossinsky, A.S.; Buttle, K.F.; Pluta, R.; Mossakowski, M.J.; Wiśniewski, H.M. Immunoultrastructural expression of intercellular adhesion molecule-1 in endothelial cell vesiculotubular structures and vesiculovacuolar organelles in blood-brain barrier development and injury. Cell Tissue Res. 1999, 295, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, B.; Ransohoff, R.M. Capture, crawl, cross: The T cell code to breach the blood-brain barriers. Trends Immunol. 2012, 33, 579–589. [Google Scholar] [CrossRef]
- Carman, C.V.; Springer, T.A. A transmigratory cup in leukocyte diapedesis both through individual vascular endothelial cells and between them. J. Cell Biol. 2004, 167, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Froio, R.M.; Sciuto, T.E.; Dvorak, A.M.; Alon, R.; Luscinskas, F.W. ICAM-1 regulates neutrophil adhesion and transcellular migration of TNF-alpha-activated vascular endothelium under flow. Blood 2005, 106, 584–592. [Google Scholar] [CrossRef] [Green Version]
- Ghaffarian, R.; Bhowmick, T.; Muro, S. Transport of nanocarriers across gastrointestinal epithelial cells by a new transcellular route induced by targeting ICAM-1. J. Control. Release 2012, 163, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Hsu, J.; Rappaport, J.; Muro, S. Specific binding, uptake, and transport of ICAM-1-targeted nanocarriers across endothelial and subendothelial cell components of the blood-brain barrier. Pharm. Res. 2014, 31, 1855–1866. [Google Scholar] [CrossRef] [Green Version]
- Manthe, R.L.; Loeck, M.; Bhowmick, T.; Solomon, M.; Muro, S. Intertwined mechanisms define transport of anti-ICAM nanocarriers across the endothelium and brain delivery of a therapeutic enzyme. J. Control. Release 2020, 324, 181–193. [Google Scholar] [CrossRef]
- Staunton, D.E.; Dustin, M.L.; Springer, T.A. Functional cloning of ICAM-2, a cell adhesion ligand for LFA-1 homologous to ICAM-1. Nature 1989, 339, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Dorovini-Zis, K. Upregulation of intercellular adhesion molecule-1 (ICAM-1) expression in primary cultures of human brain microvessel endothelial cells by cytokines and lipopolysaccharide. J. Neuroimmunol. 1992, 39, 11–21. [Google Scholar] [CrossRef]
- Sato, K.; Tachikawa, M.; Watanabe, M.; Uchida, Y.; Terasaki, T. Selective Protein Expression Changes of Leukocyte-Migration-Associated Cluster of Differentiation Antigens at the Blood-Brain Barrier in a Lipopolysaccharide-Induced Systemic Inflammation Mouse Model without Alteration of Transporters, Receptors or Tight Junction-Related Protein. Biol. Pharm. Bull. 2019, 42, 944–953. [Google Scholar] [PubMed] [Green Version]
- Connolly, E.S., Jr.; Winfree, C.J.; Springer, T.A.; Naka, Y.; Liao, H.; Yan, S.D.; Stern, D.M.; Solomon, R.A.; Gutierrez-Ramos, J.C.; Pinsky, D.J. Cerebral protection in homozygous null ICAM-1 mice after middle cerebral artery occlusion. Role of neutrophil adhesion in the pathogenesis of stroke. J. Clin. Investig. 1996, 97, 209–216. [Google Scholar] [CrossRef]
- McFarland, H.F.; Martin, R. Multiple sclerosis: A complicated picture of autoimmunity. Nat. Immunol. 2007, 8, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Lossinsky, A.S.; Mossakowski, M.J.; Pluta, R.; Wisniewski, H.M. Intercellular adhesion molecule-1 (ICAM-1) upregulation in human brain tumors as an expression of increased blood-brain barrier permeability. Brain Pathol. 1995, 5, 339–344. [Google Scholar] [CrossRef]
- Mrozowska, P.S.; Fukuda, M. Regulation of podocalyxin trafficking by Rab small GTPases in epithelial cells. Small GTPases 2016, 7, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.N.; Schnaar, R.L.; Konstantopoulos, K. Podocalyxin-like protein is an E-/L-selectin ligand on colon carcinoma cells: Comparative biochemical properties of selectin ligands in host and tumor cells. Am. J. Physiol. Cell Physiol. 2009, 296, C505–C513. [Google Scholar] [CrossRef]
- Agarwal, N.; Lippmann, E.S.; Shusta, E.V. Identification and expression profiling of blood-brain barrier membrane proteins. J. Neurochem. 2010, 112, 625–635. [Google Scholar] [CrossRef] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ito, S.; Oishi, M.; Ogata, S.; Uemura, T.; Couraud, P.-O.; Masuda, T.; Ohtsuki, S. Identification of Cell-Surface Proteins Endocytosed by Human Brain Microvascular Endothelial Cells In Vitro. Pharmaceutics 2020, 12, 579. https://doi.org/10.3390/pharmaceutics12060579
Ito S, Oishi M, Ogata S, Uemura T, Couraud P-O, Masuda T, Ohtsuki S. Identification of Cell-Surface Proteins Endocytosed by Human Brain Microvascular Endothelial Cells In Vitro. Pharmaceutics. 2020; 12(6):579. https://doi.org/10.3390/pharmaceutics12060579
Chicago/Turabian StyleIto, Shingo, Mariko Oishi, Seiryo Ogata, Tatsuki Uemura, Pierre-Olivier Couraud, Takeshi Masuda, and Sumio Ohtsuki. 2020. "Identification of Cell-Surface Proteins Endocytosed by Human Brain Microvascular Endothelial Cells In Vitro" Pharmaceutics 12, no. 6: 579. https://doi.org/10.3390/pharmaceutics12060579