Effects of Decomplexation Rates on Ternary Gene Complex Transfection with α-Poly(l-Lysine) or ε-Poly(l-Lysine) as a Decomplexation Controller in An Easy-To-Transfect Cell or A Hard-To-Transfect Cell
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Cell Culture
2.2. Preparation of the Nanocomplex
2.3. Particle Size and Zeta Potential of Nanocomplexes
2.4. Dye Quenching-Based pDNA Compactness Assay
2.5. Heparin-Induced Decomplexation of the Nanocomplexes
2.6. Transfection Efficiency and Cytotoxicity of the Nanocomplex
2.7. Proton Buffering Capacity of the Mixtures of bPEI and PL
2.8. Cellular Uptake and Nuclear Uptake of the Nanocomplexes
2.9. Intracellular Localization of the Nanocomplexes
2.10. Statistical Analysis
3. Results and Discussion
3.1. Decomplexation of the bPEI-PL/pDNA Nanocomplex
3.2. Physicochemical Characteristics of the bPEI-PL/pDNA Nanocomplexes
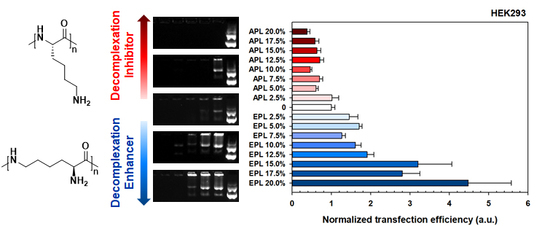
3.3. Transfection Efficiency and Cytotoxicity of the bPEI-PL/pDNA Nanocomplex
3.4. Proton Buffering Capacities of the Mixtures of bPEI and PL for Estimating Endosomal Escape of Polyplex
3.5. Cellular Uptake, Nuclear Uptake, and Subcellular Localization of the bPEI-PL/pDNA Nanocomplex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Salameh, J.W.; Zhou, L.; Ward, S.M.; Santa Chalarca, C.F.; Emrick, T.; Figueiredo, M.L. Polymer-mediated gene therapy: Recent advances and merging of delivery techniques. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1598. [Google Scholar] [CrossRef]
- Chen, J.; Wang, K.; Wu, J.; Tian, H.; Chen, X. Polycations for Gene Delivery: Dilemmas and Solutions. Bioconjug. Chem. 2019, 30, 338–349. [Google Scholar] [CrossRef]
- Keles, E.; Song, Y.; Du, D.; Dong, W.J.; Lin, Y. Recent progress in nanomaterials for gene delivery applications. Biomater. Sci. 2016, 4, 1291–1309. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Huh, K.M.; Bae, Y.H. Polymeric nucleic acid carriers: Current issues and novel design approaches. J. Control Release 2012, 164, 256–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigsby, C.L.; Leong, K.W. Balancing protection and release of DNA: Tools to address a bottleneck of non-viral gene delivery. J. R. Soc. Interface 2010, 7 (Suppl. 1), S67–S82. [Google Scholar] [CrossRef]
- Arigita, C.; Zuidam, N.J.; Crommelin, D.J.; Hennink, W.E. Association and dissociation characteristics of polymer/DNA complexes used for gene delivery. Pharm. Res. 1999, 16, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Hwang, H.S.; Shim, M.S.; Cho, Y.Y.; Lee, J.Y.; Lee, H.S.; Kang, H.C. Controlling complexation/decomplexation and sizes of polymer-based electrostatic pDNA polyplexes is one of the key factors in effective transfection. Colloids Surf. B Biointerfaces 2019, 184, 110497. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.S.; Kang, H.C.; Bae, Y.H. Bioreducible polymers as a determining factor for polyplex decomplexation rate and transfection. Biomacromolecules 2013, 14, 548–556. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.C.; Lee, M.; Bae, Y.H. Polymeric gene carriers. Crit. Rev. Eukaryot Gene Expr. 2005, 15, 317–342. [Google Scholar] [CrossRef] [PubMed]
- Pollard, H.; Toumaniantz, G.; Amos, J.L.; Avet-Loiseau, H.; Guihard, G.; Behr, J.P.; Escande, D. Ca2+-sensitive cytosolic nucleases prevent efficient delivery to the nucleus of injected plasmids. J. Gene Med. 2001, 3, 153–164. [Google Scholar] [CrossRef]
- Lechardeur, D.; Sohn, K.J.; Haardt, M.; Joshi, P.B.; Monck, M.; Graham, R.W.; Beatty, B.; Squire, J.; O’Brodovich, H.; Lukacs, G.L. Metabolic instability of plasmid DNA in the cytosol: A potential barrier to gene transfer. Gene Ther. 1999, 6, 482–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.J.; Kim, S.W. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm. Res. 2009, 26, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, G.L.; Haggie, P.; Seksek, O.; Lechardeur, D.; Freedman, N.; Verkman, A.S. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 2000, 275, 1625–1629. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.L.; Davies, M.C.; Rackstraw, B.J.; Roberts, C.J.; Stolnik, S.; Tendler, S.J.; Williams, P.M. Observation of DNA-polymer condensate formation in real time at a molecular level. FEBS Lett. 2000, 480, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Vacik, J.; Dean, B.S.; Zimmer, W.E.; Dean, D.A. Cell-specific nuclear import of plasmid DNA. Gene Ther. 1999, 6, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, D.V.; Fidelman, N.A.; Dan, N.; Lauffenburger, D.A. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 2000, 67, 598–606. [Google Scholar] [CrossRef]
- Kang, H.C.; Kang, H.J.; Bae, Y.H. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials 2011, 32, 1193–1203. [Google Scholar] [CrossRef] [Green Version]
- Miyata, K.; Kakizawa, Y.; Nishiyama, N.; Harada, A.; Yamasaki, Y.; Koyama, H.; Kataoka, K. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J. Am. Chem. Soc. 2004, 126, 2355–2361. [Google Scholar] [CrossRef]
- Mann, A.; Thakur, G.; Shukla, V.; Singh, A.K.; Khanduri, R.; Naik, R.; Jiang, Y.; Kalra, N.; Dwarakanath, B.S.; Langel, U.; et al. Differences in DNA condensation and release by lysine and arginine homopeptides govern their DNA delivery efficiencies. Mol. Pharm. 2011, 8, 1729–1741. [Google Scholar] [CrossRef]
- Choi, J.Y.; Ryu, K.; Lee, G.J.; Kim, K.; Kim, T.I. Agmatine-Containing Bioreducible Polymer for Gene Delivery Systems and Its Dual Degradation Behavior. Biomacromolecules 2015, 16, 2715–2725. [Google Scholar] [CrossRef]
- Kim, Y.H.; Park, J.H.; Lee, M.; Kim, Y.H.; Park, T.G.; Kim, S.W. Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J. Control. Release 2005, 103, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Gabrielson, N.P.; Pack, D.W. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules 2006, 7, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Bae, Y.H. Co-delivery of small interfering RNA and plasmid DNA using a polymeric vector incorporating endosomolytic oligomeric sulfonamide. Biomaterials 2011, 32, 4914–4924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Ryu, K.; Choi, Y.S.; Cho, Y.Y.; Lee, J.Y.; Lee, H.S.; Kang, H.C. Effects of the Physicochemical, Colloidal, and Biological Characteristics of Different Polymer Structures between alpha-Poly(l-lysine) and epsilon-Poly(l-lysine) on Polymeric Gene Delivery. Biomacromolecules 2018, 19, 2483–2495. [Google Scholar] [CrossRef]
- Guo, J.; Wei, Y.; Zhou, D.; Cai, P.; Jing, X.; Chen, X.S.; Huang, Y. Chemosynthesis of poly(epsilon-lysine)-analogous polymers by microwave-assisted click polymerization. Biomacromolecules 2011, 12, 737–746. [Google Scholar] [CrossRef]
- Wang, T.; Larcher, L.M.; Ma, L.; Veedu, R.N. Systematic Screening of Commonly Used Commercial Transfection Reagents towards Efficient Transfection of Single-Stranded Oligonucleotides. Molecules 2018, 23, 2564. [Google Scholar] [CrossRef] [Green Version]
- Goyal, R.; Tripathi, S.K.; Tyagi, S.; Sharma, A.; Ram, K.R.; Chowdhuri, D.K.; Shukla, Y.; Kumar, P.; Gupta, K.C. Linear PEI nanoparticles: Efficient pDNA/siRNA carriers in vitro and in vivo. Nanomedicine 2012, 8, 167–175. [Google Scholar] [CrossRef]
- Das, J.; Han, J.W.; Choi, Y.J.; Song, H.; Cho, S.G.; Park, C.; Seo, H.G.; Kim, J.H. Cationic lipid-nanoceria hybrids, a novel nonviral vector-mediated gene delivery into mammalian cells: Investigation of the cellular uptake mechanism. Sci. Rep. 2016, 6, 29197. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.; Smart, T.G. HEK293 cell line: A vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods 2005, 51, 187–200. [Google Scholar] [CrossRef]
- Chin, C.L.; Goh, J.B.; Srinivasan, H.; Liu, K.I.; Gowher, A.; Shanmugam, R.; Lim, H.L.; Choo, M.; Tang, W.Q.; Tan, A.H.; et al. A human expression system based on HEK293 for the stable production of recombinant erythropoietin. Sci. Rep. 2019, 9, 16768. [Google Scholar] [CrossRef] [Green Version]
- Donato, M.T.; Lahoz, A.; Castell, J.V.; Gomez-Lechon, M.J. Cell lines: A tool for in vitro drug metabolism studies. Curr. Drug Metab. 2008, 9, 1–11. [Google Scholar] [PubMed]
- Choi, J.M.; Oh, S.J.; Lee, S.Y.; Im, J.H.; Oh, J.M.; Ryu, C.S.; Kwak, H.C.; Lee, J.Y.; Kang, K.W.; Kim, S.K. HepG2 cells as an in vitro model for evaluation of cytochrome P450 induction by xenobiotics. Arch. Pharm. Res. 2015, 38, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Chen, H.; Jin, R.; Weng, T.; Ho, J.K.; You, C.; Zhang, L.; Wang, X.; Han, C. Non-viral gene delivery systems for tissue repair and regeneration. J. Transl. Med. 2018, 16, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ooi, A.; Wong, A.; Esau, L.; Lemtiri-Chlieh, F.; Gehring, C. A Guide to Transient Expression of Membrane Proteins in HEK-293 Cells for Functional Characterization. Front. Physiol. 2016, 7, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Intra, J.; Salem, A.K. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. J. Control. Release 2008, 130, 129–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]












© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Ryu, K.; Cho, H.; Shim, M.S.; Cho, Y.-Y.; Lee, J.Y.; Lee, H.S.; Kang, H.C. Effects of Decomplexation Rates on Ternary Gene Complex Transfection with α-Poly(l-Lysine) or ε-Poly(l-Lysine) as a Decomplexation Controller in An Easy-To-Transfect Cell or A Hard-To-Transfect Cell. Pharmaceutics 2020, 12, 490. https://doi.org/10.3390/pharmaceutics12060490
Kim K, Ryu K, Cho H, Shim MS, Cho Y-Y, Lee JY, Lee HS, Kang HC. Effects of Decomplexation Rates on Ternary Gene Complex Transfection with α-Poly(l-Lysine) or ε-Poly(l-Lysine) as a Decomplexation Controller in An Easy-To-Transfect Cell or A Hard-To-Transfect Cell. Pharmaceutics. 2020; 12(6):490. https://doi.org/10.3390/pharmaceutics12060490
Chicago/Turabian StyleKim, Kyoungnam, Kitae Ryu, Hana Cho, Min Suk Shim, Yong-Yeon Cho, Joo Young Lee, Hye Suk Lee, and Han Chang Kang. 2020. "Effects of Decomplexation Rates on Ternary Gene Complex Transfection with α-Poly(l-Lysine) or ε-Poly(l-Lysine) as a Decomplexation Controller in An Easy-To-Transfect Cell or A Hard-To-Transfect Cell" Pharmaceutics 12, no. 6: 490. https://doi.org/10.3390/pharmaceutics12060490