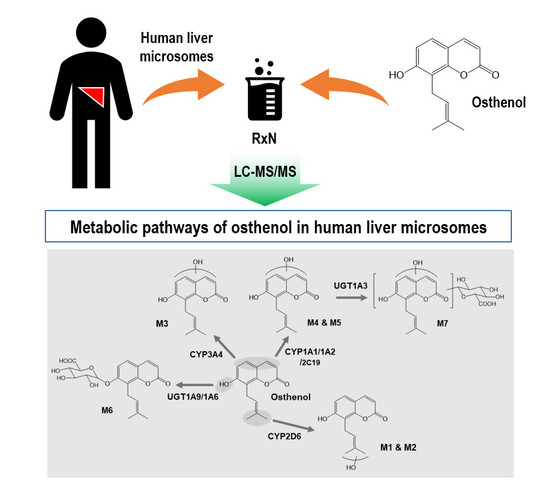
Characterization of CYPs and UGTs Involved in Human Liver Microsomal Metabolism of Osthenol
Abstract
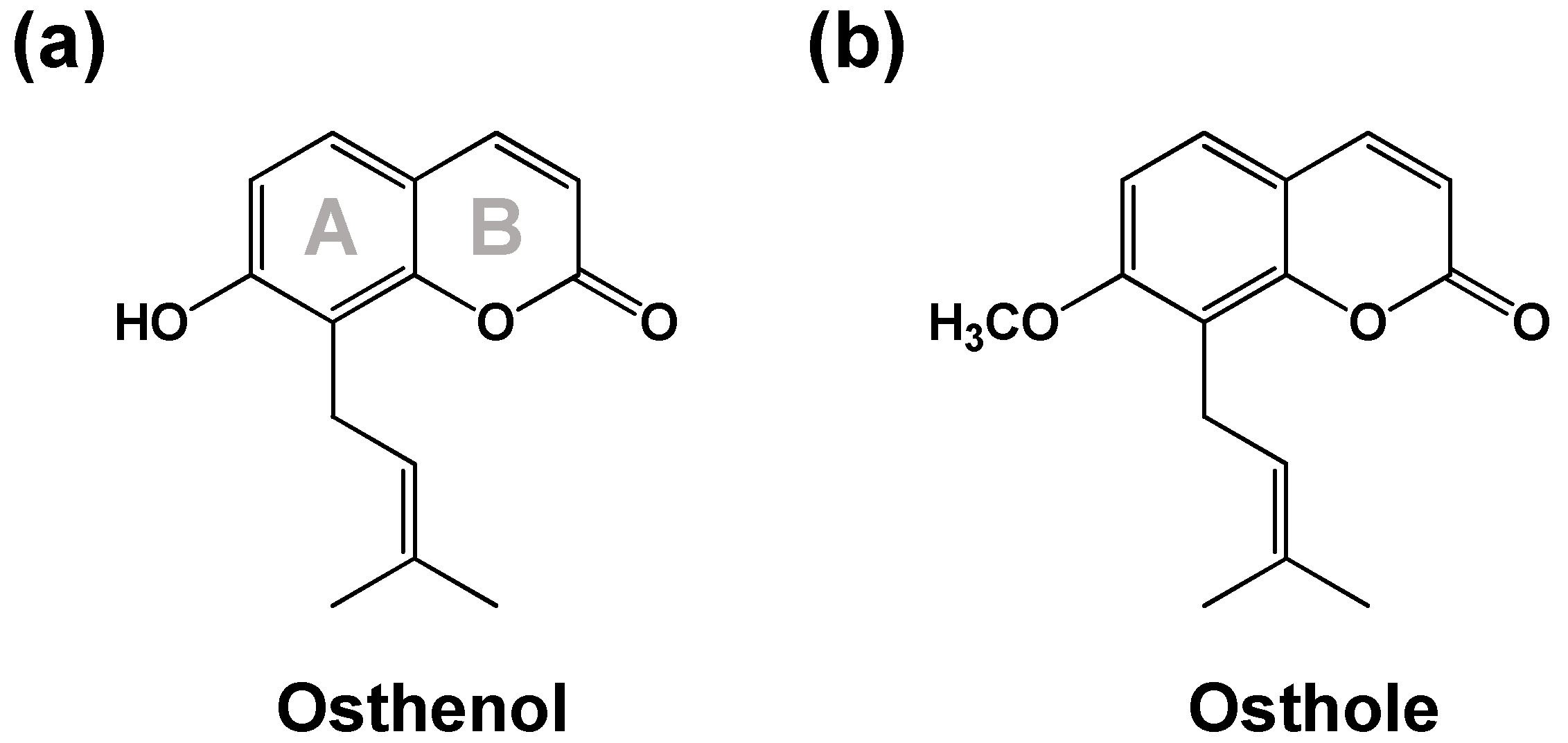
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Identification of Osthenol Metabolites in Human Liver Microsomes
2.3. Metabolism of Osthenol in Human Recombinant cDNA-Expressed CYP and UGT Isoforms
2.4. Reaction Phenotyping for M7
2.5. Instruments
3. Results
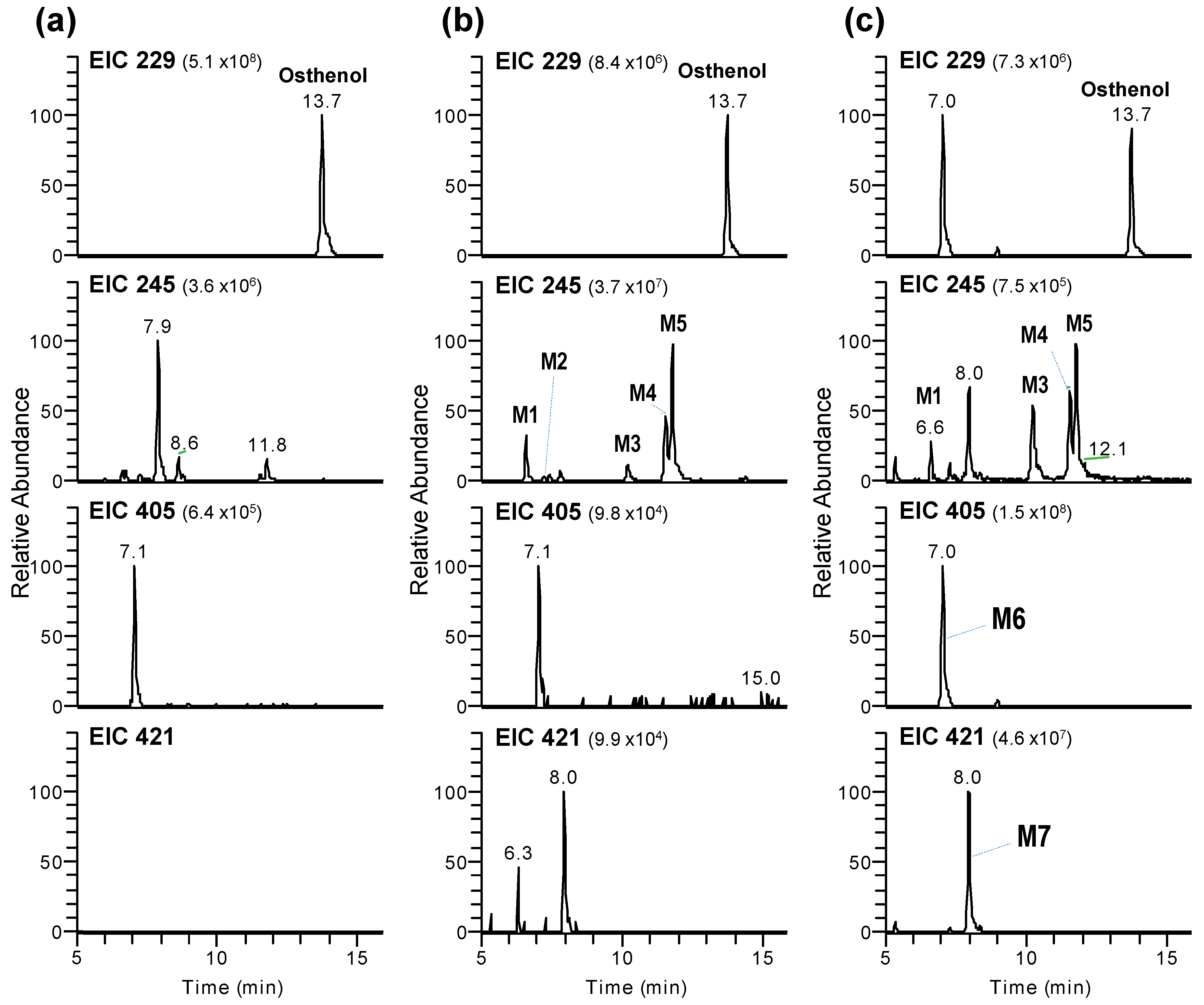
3.1. Identification of Phase I and Phase II Metabolites of Osthenol in HLMs
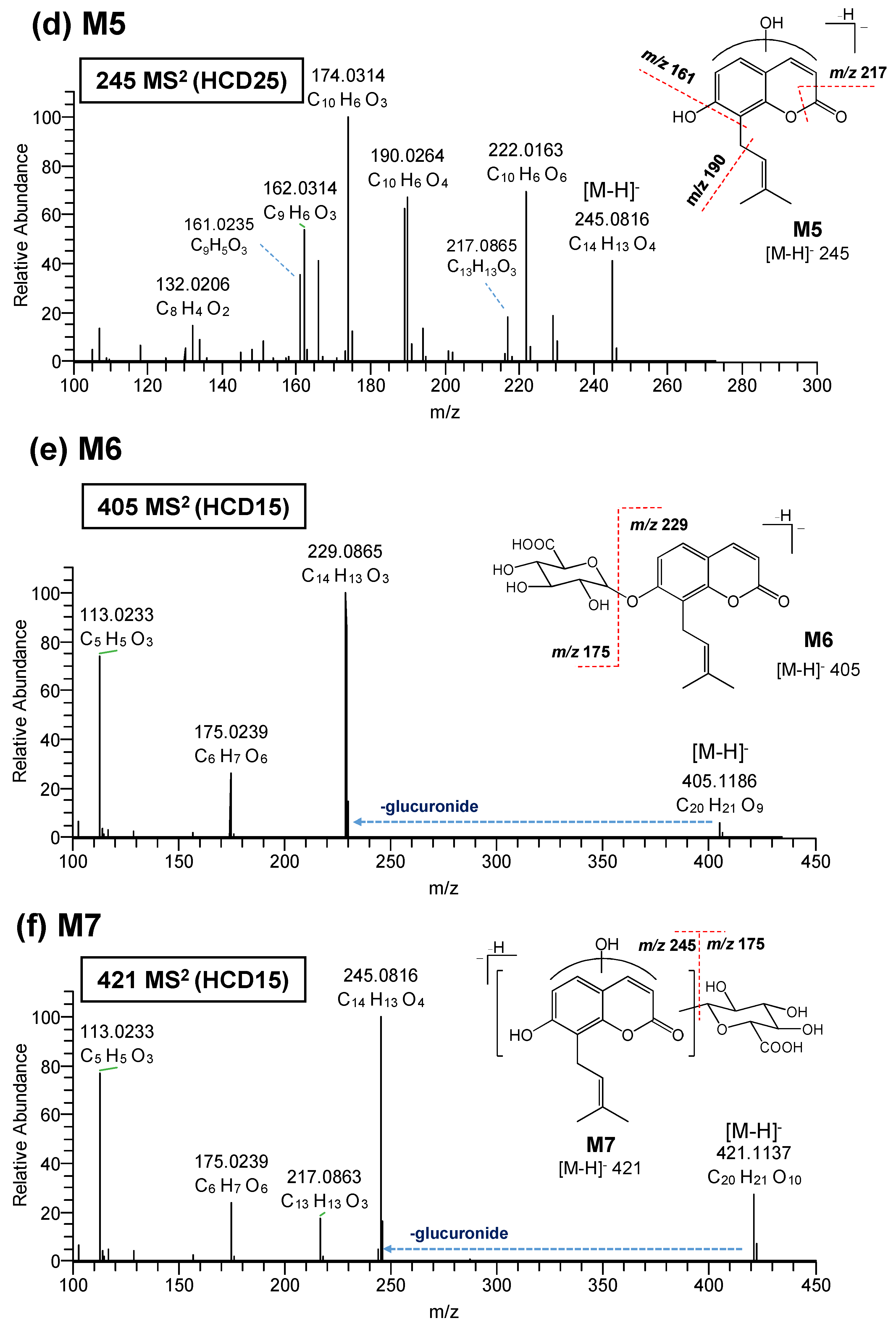
3.2. Elucidation of Metabolite Structure
3.3. Reaction Phenotyping of Osthenol Metabolism Using cDNA-Expressed Recombinant CYP and UGT Isoforms
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Seo, E.K.; Kim, K.H.; Kim, M.K.; Cho, M.H.; Choi, E.; Kim, K.; Mar, W. Inhibitors of 5alpha -reductase type I in LNCaP cells from the roots of Angelica koreana. Planta Med. 2002, 68, 162–163. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, G.; Chen, B.; Xie, Y.; Wu, H.; Wu, Y.; Yan, C.; Wang, J. Separation and quantitative analysis of coumarin compounds from Angelica dahurica (Fisch. ex Hoffm) Benth. et Hook. f by pressurized capillary electrochromatography. J. Pharm. Biomed. Anal. 2006, 41, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Karamat, F.; Olry, A.; Munakata, R.; Koeduka, T.; Sugiyama, A.; Paris, C.; Hehn, A.; Bourgaud, F.; Yazaki, K. A coumarin-specific prenyltransferase catalyzes the crucial biosynthetic reaction for furanocoumarin formation in parsley. Plant J. 2014, 77, 627–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, C.; Itoigawa, M.; Ju-ichi, M.; Sakamoto, N.; Tokuda, H.; Nishino, H.; Furukawa, H. Antitumor-promoting activity of coumarins from citrus plants. Planta Med. 2005, 71, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Montagner, C.; de Souza, S.M.; Groposoa, C.; Delle Monache, F.; Smania, E.F.; Smania, A., Jr. Antifungal activity of coumarins. Z. Naturforsch. C 2008, 63, 21–28. [Google Scholar] [CrossRef] [PubMed]
- de Souza, S.M.; Delle Monache, F.; Smania, A., Jr. Antibacterial activity of coumarins. Z. Naturforsch. C 2005, 60, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Zschocke, S.; Reininger, E.; Bauer, R. Inhibitory effects of Angelica pubescens f. biserrata on 5-lipoxygenase and cyclooxygenase. Planta Med. 1998, 64, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.R.; Leung, W.N.; Cheung, H.Y.; Chan, C.W. Osthole: A review on its bioactivities, pharmacological properties, and potential as alternative medicine. Evid. Based Complement. Altern. Med. 2015, 2015, 919616. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Xu, H.; Wang, K.; Zhao, Z.; Hu, M. Determination of osthol and its metabolites in a phase I reaction system and the Caco-2 cell model by HPLC-UV and LC-MS/MS. J. Pharm. Biomed. Anal. 2009, 49, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Wang, C.Y.; Hou, J.; Zhang, B.J.; Deng, S.; Tian, Y.; Huang, S.S.; Zhang, H.L.; Shu, X.H.; Zhen, Y.H.; et al. Isolation and identification of metabolites of osthole in rats. Xenobiotica 2012, 42, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Cho, P.J.; Nam, W.; Lee, D.; Lee, T.; Lee, S. Selective inhibitory effect of osthenol on human cytochrome 2C8. Bull. Korean Chem. Soc. 2018, 39, 801–805. [Google Scholar] [CrossRef]
- Li, J.; Chan, W. Investigation of the biotransformation of osthole by liquid chromatography/tandem mass spectrometry. J. Pharm. Biomed. Anal. 2013, 74, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Liu, J.P.; Lai, X.S. Substrate specificity, inhibitors and regulation of human cytochrome P450 2D6 and implications in drug development. Curr. Med. Chem. 2009, 16, 2661–2805. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.F.; Dickins, M. Substrate SARs in human P450s. Drug Discov. Today 2002, 7, 918–925. [Google Scholar] [CrossRef]
- Docampo, M.; Olubu, A.; Wang, X.; Pasinetti, G.; Dixon, R.A. Glucuronidated flavonoids in neurological protection: Structural analysis and approaches for chemical and biological synthesis. J. Agric. Food Chem. 2017, 65, 7607–7623. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Kulkarni, K.; Basu, S.; Zhang, S.; Hu, M. First-pass metabolism via UDP-glucuronosyltransferase: A barrier to oral bioavailability of phenolics. J. Pharm. Sci. 2011, 100, 3655–3681. [Google Scholar] [CrossRef] [PubMed]
- Uchaipichat, V.; Mackenzie, P.I.; Guo, X.H.; Gardner-Stephen, D.; Galetin, A.; Houston, J.B.; Miners, J.O. Human udp-glucuronosyltransferases: Isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclofenac and probenecid. Drug Metab. Dispos. 2004, 32, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.C.; Ge, G.B.; Liu, H.X.; Zhang, Y.Y.; Wang, L.M.; Zhang, J.W.; Yin, L.; Li, W.; Fang, Z.Z.; Wu, J.J.; et al. Identification and characterization of human UDP-glucuronosyltransferases responsible for the in vitro glucuronidation of daphnetin. Drug Metab. Dispos. 2010, 38, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Luukkanen, L.; Taskinen, J.; Kurkela, M.; Kostiainen, R.; Hirvonen, J.; Finel, M. Kinetic characterization of the 1A subfamily of recombinant human UDP-glucuronosyltransferases. Drug Metab. Dispos. 2005, 33, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
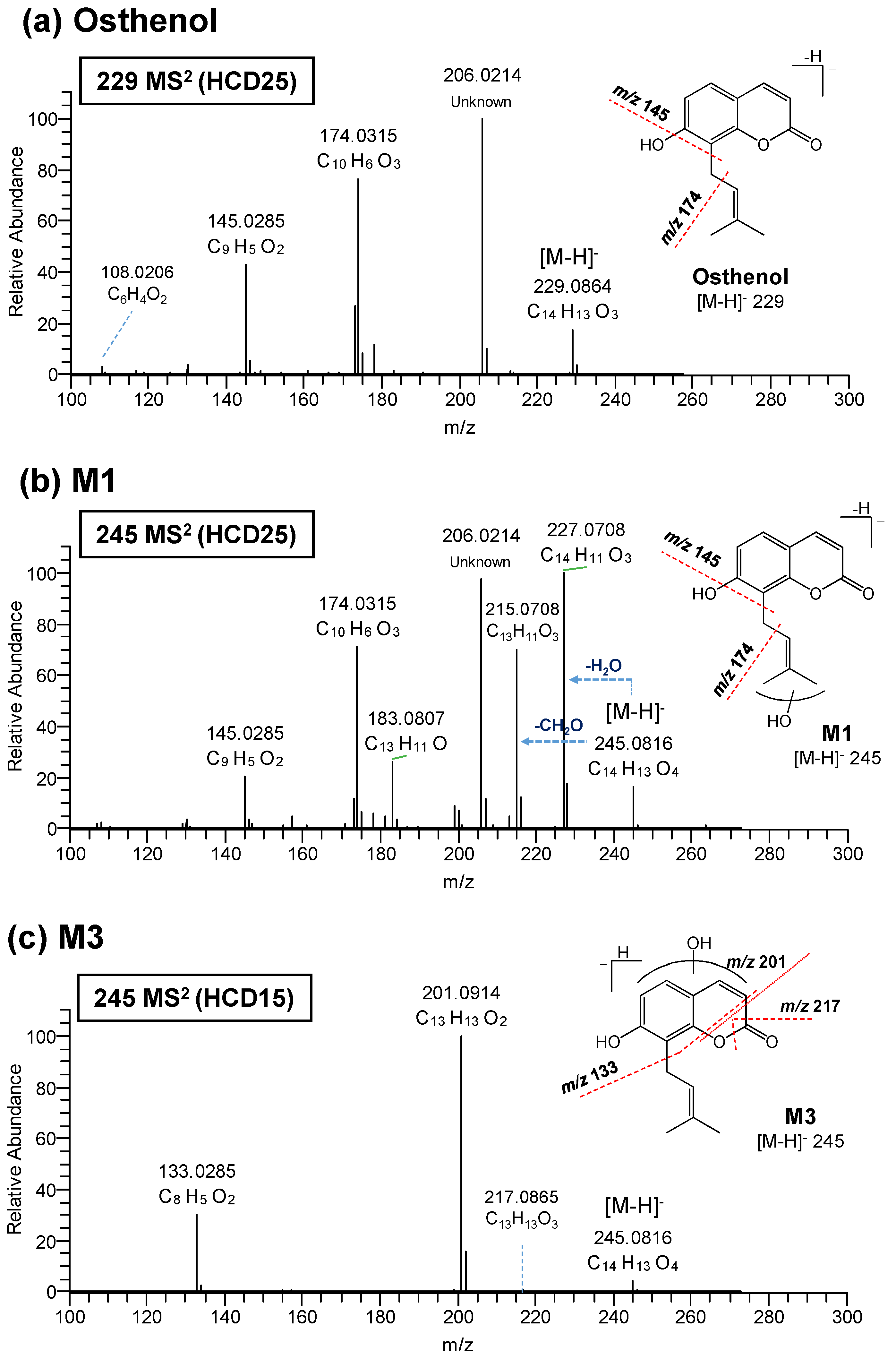



| Compound | Precursor Ions (m/z) | HCD (eV) | Product Ion (m/z) | Elemental Comp. (exp.) | Error (ppm) | Mass Shift (Da) | ||
|---|---|---|---|---|---|---|---|---|
| MS2 | Elemental Comp. (exp.) | Error (ppm) | ||||||
| Osthenol | 229.0864 | C14H13O3 | −0.5 | 25 | 206.0214 | Unknown | NA | |
| 25 | 174.0315 | C10H6O3 | −1.1 | |||||
| 25 | 145.0285 | C9H5O2 | −3.2 | |||||
| 25 | 130.0416 | C9H6O | −1.8 | |||||
| 25 | 108.0206 | C6H4O2 | −5.0 | |||||
| M1 | 245.0816 | C14H13O4 | 1.0 | 25 | 227.0708 | C14H11O3 | −0.1 | -H2O |
| 25 | 215.0708 | C13H11O3 | −0.1 | -CH2O | ||||
| 25 | 206.0214 | Unknown | NA | |||||
| 25 | 183.0807 | C13H11O | −1.4 | |||||
| 25 | 174.0315 | C10H6O3 | −1.3 | |||||
| 25 | 145.0285 | C9H5O2 | −2.9 | |||||
| 25 | 108.0207 | C6H4O2 | −4.2 | |||||
| M2 | 245.0816 | C14H13O4 | 2.9 | 25 | 227.0708 | C14H11O3 | 2.7 | -H2O |
| 25 | 215.0707 | C13H11O3 | 2.2 | |||||
| 25 | 206.0214 | Unknown | NA | |||||
| 25 | 183.0808 | C13H11O | 1.9 | |||||
| 25 | 174.0314 | C10H6O3 | 1.5 | |||||
| 25 | 145.0285 | C9H5O2 | 0.9 | |||||
| M3 | 245.0816 | C14H13O4 | 3.2 | 15 | 217.0865 | C13H13O3 | 2.7 | |
| 15 | 201.0914 | C13H13O2 | 2.0 | |||||
| 15 | 133.0285 | C8H5O2 | 0.5 | |||||
| M4 | 245.0817 | C14H13O4 | 3.4 | 25 | 229.0501 | C13H9O4 | 2.5 | |
| 25 | 222.0164 | C10H6O6 | 2.3 | 206 + 16 | ||||
| 25 | 201.0914 | C13H13O2 | 1.9 | |||||
| 25 | 190.0264 | C10H6O4 | 1.8 | 174 + 16 | ||||
| 25 | 173.0965 | C12H13O | 2.4 | |||||
| 25 | 162.0314 | C9H6O3 | 1.4 | |||||
| 25 | 132.0207 | C8H4O2 | 0.9 | |||||
| M5 | 245.0816 | C14H13O4 | 1.0 | 25 | 229.0501 | C13H9O4 | 0.2 | 213 + 16 |
| 25 | 222.0163 | C10H6O6 | −0.4 | 206 + 16 | ||||
| 25 | 217.0865 | C13H13O3 | 0.4 | |||||
| 25 | 190.0264 | C10H6O4 | −1.4 | 174 + 16 | ||||
| 25 | 174.0314 | C10H6O3 | −1.6 | |||||
| 25 | 166.0262 | C8H6O4 | −2.3 | |||||
| 25 | 162.0314 | C9H6O3 | −1.5 | |||||
| 25 | 161.0235 | C9H5O3 | −2.6 | 145 + 16 | ||||
| 25 | 132.0206 | C8H4O2 | −3.7 | |||||
| M6 | 405.1186 | C20H21O9 | 1.5 | 15 | 229.0865 | C14H13O3 | 2.7 | |
| 15 | 175.0239 | C6H7O6 | 1.3 | |||||
| 15 | 113.0233 | C5H5O3 | −0.1 | |||||
| M7 | 421.1137 | C20H21O10 | 1.8 | 15 | 245.0816 | C14H13O4 | 3.1 | |
| 15 | 217.0863 | C13H13O3 | 1.8 | |||||
| 15 | 175.0239 | C6H7O6 | 1.3 | |||||
| 15 | 113.2333 | C5H5O3 | −0.2 | |||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, P.J.; Paudel, S.; Lee, D.; Jin, Y.J.; Jo, G.; Jeong, T.C.; Lee, S.; Lee, T. Characterization of CYPs and UGTs Involved in Human Liver Microsomal Metabolism of Osthenol. Pharmaceutics 2018, 10, 141. https://doi.org/10.3390/pharmaceutics10030141
Cho PJ, Paudel S, Lee D, Jin YJ, Jo G, Jeong TC, Lee S, Lee T. Characterization of CYPs and UGTs Involved in Human Liver Microsomal Metabolism of Osthenol. Pharmaceutics. 2018; 10(3):141. https://doi.org/10.3390/pharmaceutics10030141
Chicago/Turabian StyleCho, Pil Joung, Sanjita Paudel, Doohyun Lee, Yun Ji Jin, GeunHyung Jo, Tae Cheon Jeong, Sangkyu Lee, and Taeho Lee. 2018. "Characterization of CYPs and UGTs Involved in Human Liver Microsomal Metabolism of Osthenol" Pharmaceutics 10, no. 3: 141. https://doi.org/10.3390/pharmaceutics10030141