Whole Genomic Analysis of Human G1P[8] Rotavirus Strains From Different Age Groups in China
Abstract
:1. Introduction
2. Results, Discussion and Conclusion
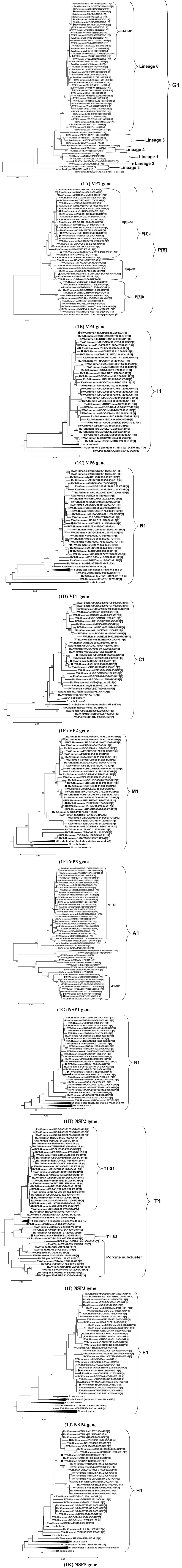
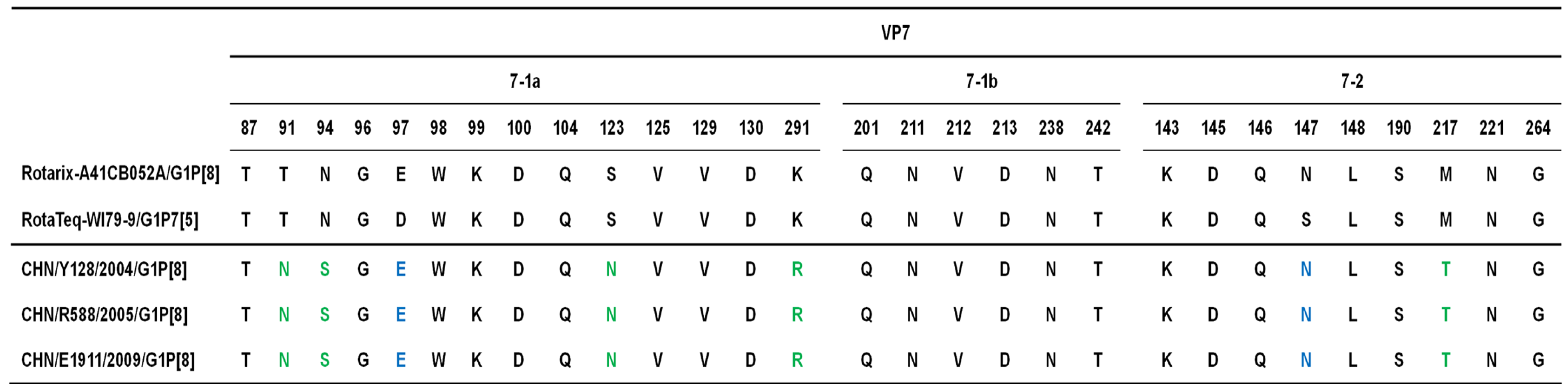
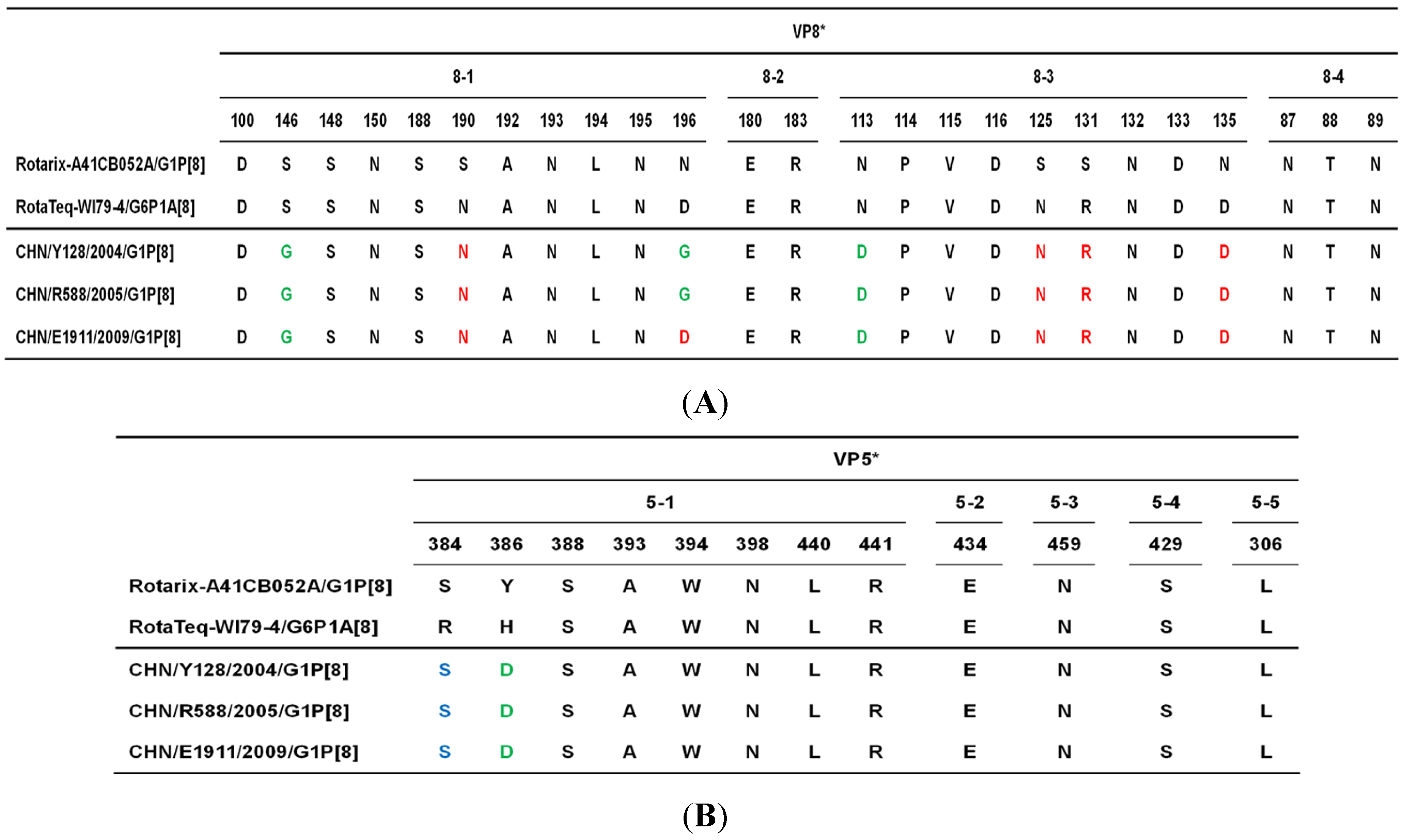
3. Materials and Methods
3.1. Virus Strains
| RVA strain | Age and sex of patient | Duration of diarrhea | Clinical signs | Duration of Hospitalization |
|---|---|---|---|---|
| E1911 | 8-month-old male infant | 1 day | Passing liquid stools three times a day. Mild dehydration. No vomiting or fever. | None. Treated at outpatient department. |
| R588 | 3-year-old male child | 2 days | Passing liquid stools five times a day. Mild dehydration. No vomiting or fever. | None. Treated at outpatient department. |
| Y128 | 66-year-old man | 1 day | Passing liquid stools seven times a day. Vomiting three times a day. Severe dehydration. No fever. | None. Treated at outpatient department. |
3.2. RT-PCR and Nucleotide Sequencing
3.3. Sequence Analyses
3.4. Nucleotide Sequence Accession Numbers
Supplementary Files
Acknowledgments
Conflict of Interest
References
- Estes, M.K.; Kapikian, A.Z. Rotaviruses. In Fields Virology, 5th; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Straus, S.E., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 1917–1974. [Google Scholar]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Banyai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gomara, M.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar]
- Matthijnssens, J.; Bilcke, J.; Ciarlet, M.; Martella, V.; Bányai, K.; Rahman, M.; Zeller, M.; Beutels, P.; van Damme, P.; van Ranst, M. Rotavirus disease and vaccination: Impact on genotype diversity. Future Microbiol. 2009, 4, 1303–1316. [Google Scholar] [CrossRef]
- Santos, N.; Hoshino, Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 2005, 15, 29–56. [Google Scholar] [CrossRef]
- Jiang, V.; Jiang, B.; Tate, J.; Parashar, U.D.; Patel, M.M. Performance of rotavirus vaccines in developed and developing countries. Hum. Vaccin. 2010, 6, 532–542. [Google Scholar] [CrossRef]
- Jin, H.; Wang, B.; Fang, Z.; Duan, Z.; Gao, Q.; Liu, N.; Zhang, L.; Qian, Y.; Gong, S.; Zhu, Q.; Shen, X.; Wu, Q. Hospital-based study of the economic burden associated with rotavirus diarrhea in eastern China. Vaccine 2011, 29, 7801–7806. [Google Scholar] [CrossRef]
- Kawai, K.; O'Brien, M.A.; Goveia, M.G.; Mast, T.C.; El Khoury, A.C. Burden of rotavirus gastroenteritis and distribution of rotavirus strains in Asia: A systematic review. Vaccine 2012, 30, 1244–1254. [Google Scholar]
- Wang, Y.H.; Kobayashi, N.; Zhou, D.J.; Yang, Z.Q.; Zhou, X.; Peng, J.S.; Zhu, Z.R.; Zhao, D.F.; Liu, M.Q.; Gong, J. Molecular epidemiologic analysis of group A rotaviruses in adults and children with diarrhea in Wuhan city, China, 2000-2006. Arch. Virol. 2007, 152, 669–685. [Google Scholar] [CrossRef]
- Wang, Y.H.; Zhou, X.; Ghosh, S.; Zhou, D.J.; Pang, B.B.; Peng, J.S.; Hu, Q.; Kobayashi, N. Prevalence of human rotavirus genotypes in Wuhan, China, during 2008-2011: Changing trend of predominant genotypes and emergence of strains with the P[8]b subtype of the VP4 gene. Arch. Virol. 2011, 156, 2221–2231. [Google Scholar] [CrossRef]
- Fu, C.; Tate, J.E.; Jiang, B. Effectiveness of Lanzhou lamb rotavirus vaccine against hospitalized gastroenteritis: further analysis and update. Hum. Vaccin. 2010, 6. [Google Scholar] [CrossRef]
- Ghosh, S.; Kobayashi, N. Whole-genomic analysis of rotavirus strains: Current status and future prospects. Future Microbiol. 2011, 6, 1049–1065. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Go´mara, M.I.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef]
- Ghosh, S.; Paul, S.K.; Yamamoto, D.; Nagashima, S.; Kobayashi, N. Full genomic analyses of human rotavirus strains possessing the rare P[8]b VP4 subtype. Infect. Genet. Evol. 2011, 11, 1481–1486. [Google Scholar] [CrossRef]
- Arora, R.; Chitambar, S.D. Full genomic analysis of Indian G1P[8] rotavirus strains. Infect. Genet. Evol. 2011, 11, 504–511. [Google Scholar] [CrossRef]
- Rahman, M.; Matthijnssens, J.; Saiada, F.; Hassan, Z.; Heylen, E.; Azim, T.; van Ranst, M. Complete genomic analysis of a Bangladeshi G1P[8] rotavirus strain detected in 2003 reveals a close evolutionary relationship with contemporary human Wa-like strains. Infect. Genet. Evol. 2010, 10, 746–754. [Google Scholar] [CrossRef]
- Bányai, K.; Mijatovic-Rustempasic, S.; Hull, J.J.; Esona, M.D.; Freeman, M.M.; Frace, A.M.; Bowen, M.D.; Gentsch, J.R. Sequencing and phylogenetic analysis of the coding region of six common rotavirus strains: evidence for intragenogroup reassortment among co-circulating G1P[8] and G2P[4] strains from the United States. J. Med. Virol. 2011, 83, 532–539. [Google Scholar] [CrossRef]
- Anderson, E.J.; Weber, S.G. Rotavirus infection in adults. Lancet Infect. Dis. 2004, 4, 91–99. [Google Scholar] [CrossRef]
- Paul, S.K.; Kobayashi, N.; Nagashima, S.; Ishino, M.; Watanabe, S.; Alam, M.M.; Ahmed, M.U.; Hossain, M.A.; Naik, T.N. Phylogenetic analysis of rotaviruses with genotypes G1, G2, G9 and G12 in Bangladesh: Evidence for a close relationship between rotaviruses from children and adults. Arch. Virol. 2008, 153, 1999–2012. [Google Scholar] [CrossRef]
- Nagashima, S.; Kobayashi, N.; Paul, S.K.; Alam, M.M.; Chawla-Sarkar, M.; Krishnan, T. Characterization of full-length VP4 genes of OP354-like P[8] human rotavirus strains detected in Bangladesh representing a novel P[8] subtype. Arch. Virol. 2009, 154, 1223–1231. [Google Scholar] [CrossRef]
- Aoki, S.T.; Settembre, E.C.; Trask, S.D.; Greenberg, H.B.; Harrison, S.C.; Dormitzer, P.R. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 2009, 324, 1444–1447. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Nason, E.B.; Prasad, B.V.; Harrison, S.C. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 2004, 430, 1053–1058. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Sun, Z.Y.; Wagner, G.; Harrison, S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO. J. 2002, 21, 885–897. [Google Scholar] [CrossRef]
- McDonald, S.M.; Matthijnssens, J.; McAllen, J.K.; Hine, E.; Overton, L.; Wang, S.; Lemey, P.; Zeller, M.; van Ranst, M.; Spiro, D.J.; et al. Evolutionary dynamics of human rotaviruses: Balancing reassortment with preferred genome constellations. PLoS. Pathog. 2009, 5. [Google Scholar] [CrossRef]
- Zeller, M.; Patton, J.T.; Heylen, E.; de Coster, S.; Ciarlet, M.; van Ranst, M.; Matthijnssens, J. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in belgium and rotaviruses in rotarix™ and RotaTeq™. J. Clin. Microbiol. 2011, 50, 966–976. [Google Scholar]
- Matthijnssens, J.; Joelsson, D.B.; Warakomski, D.J.; Zhou, T.; Mathis, P.K.; Van Maanen, M.H.; Ranheim, T.S.; Ciarlet, M. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology 2010, 403, 111–127. [Google Scholar] [CrossRef]
- Mukherjee, A.; Ghosh, S.; Bagchi, P.; Dutta, D.; Chattopadhyay, S.; Kobayashi, N.; Chawla-Sarkar, M. Full genomic analyses of human rotavirus G4P[4], G4P[6], G9P[19] and G10P[6] strains from North-eastern India: Evidence for interspecies transmission and complex reassortment events. Clin. Microbiol. Infect. 2011, 17, 1343–1346. [Google Scholar]
- Varghese, V.; Das, S.; Singh, N.B.; Kojima, K.; Bhattacharya, S.K.; Krishnan, T.; Kobayashi, N.; Naik, T.N. Molecular characterization of a human rotavirus reveals porcine characteristics in most of the genes including VP6 and NSP4. Arch. Virol. 2004, 149, 155–172. [Google Scholar]
- Chen, Y.; Wen, Y.; Liu, X.; Xiong, X.; Cao, Z.; Zhao, Q.; Yu, Y.; Yin, X.; Li, C.; Fan, Y. Full genomic analysis of human rotavirus strain TB-Chen isolated in China. Virology. 2008, 375, 361–373. [Google Scholar] [CrossRef]
- Wang, Y.H.; Kobayashi, N.; Nagashima, S.; Zhou, X.; Ghosh, S.; Peng, J.S.; Hu, Q.; Zhou, D.J.; Yang, Z.Q. Full genomic analysis of a porcine-bovine reassortant G4P[6] rotavirus strain R479 isolated from an infant in China. J. Med. Virol. 2010, 82, 1094–1102. [Google Scholar] [CrossRef]
- Lopman, B.A.; Payne, D.C.; Tate, J.E.; Patel, M.M.; Cortese, M.M.; Parashar, U.D. Post-licensure experience with rotavirus vaccination in high and middle income countries; 2006 to 2011. Curr. Opin. Virol. 2012. [Google Scholar] [CrossRef]
- Gurgel, R.G.; Bohland, A.; Vieira, S.C.; Oliveira, D.M.; Fontes, P.B.; Barros, V.F.; Ramos, M.F.; Dove, W.; Nakagomi, T.; Nakagomi, O.; et al. Incidence of rotavirus and all-cause diarrhea in northeast Brazil following the introduction of a national vaccination program. Gastroenterology 2009, 137, 1970–1975. [Google Scholar] [CrossRef]
- Blazevic, V.; Lappalainen, S.; Nurminen, K.; Huhti, L.; Vesikari, T. Norovirus VLPs and rotavirus VP6 protein as combined vaccine for childhood gastroenteritis. Vaccine 2011, 29, 8126–8133. [Google Scholar]
- Ghosh, S.; Adachi, N.; Gatheru, Z.; Nyangao, J.; Yamamoto, D.; Ishino, M.; Urushibara, N.; Kobayashi, N. Whole-genome analysis reveals the complex evolutionary dynamics of Kenyan G2P[4] human rotavirus strains. J. Gen. Virol. 2011, 92, 2201–2208. [Google Scholar] [CrossRef]
- Ghosh, S.; Kobayashi, N.; Nagashima, S.; Chawla-Sarkar, M.; Krishnan, T.; Ganesh, B.; Naik, T.N. Full genomic analysis and possible origin of a porcine G12 rotavirus strain RU172. Virus Genes 2010, 40, 382–388. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Available online: http://clustalw.ddbj.nig.ac.jp/ (accessed on 30 March 2012).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shintani, T.; Ghosh, S.; Wang, Y.-H.; Zhou, X.; Zhou, D.-J.; Kobayashi, N. Whole Genomic Analysis of Human G1P[8] Rotavirus Strains From Different Age Groups in China. Viruses 2012, 4, 1289-1304. https://doi.org/10.3390/v4081289
Shintani T, Ghosh S, Wang Y-H, Zhou X, Zhou D-J, Kobayashi N. Whole Genomic Analysis of Human G1P[8] Rotavirus Strains From Different Age Groups in China. Viruses. 2012; 4(8):1289-1304. https://doi.org/10.3390/v4081289
Chicago/Turabian StyleShintani, Tsuzumi, Souvik Ghosh, Yuan-Hong Wang, Xuan Zhou, Dun-Jin Zhou, and Nobumichi Kobayashi. 2012. "Whole Genomic Analysis of Human G1P[8] Rotavirus Strains From Different Age Groups in China" Viruses 4, no. 8: 1289-1304. https://doi.org/10.3390/v4081289





