A Gene Encoding Scots Pine Antimicrobial Protein Sp-AMP2 (PR-19) Confers Increased Tolerance against Botrytis cinerea in Transgenic Tobacco
Abstract
:1. Introduction
2. Materials and Methods
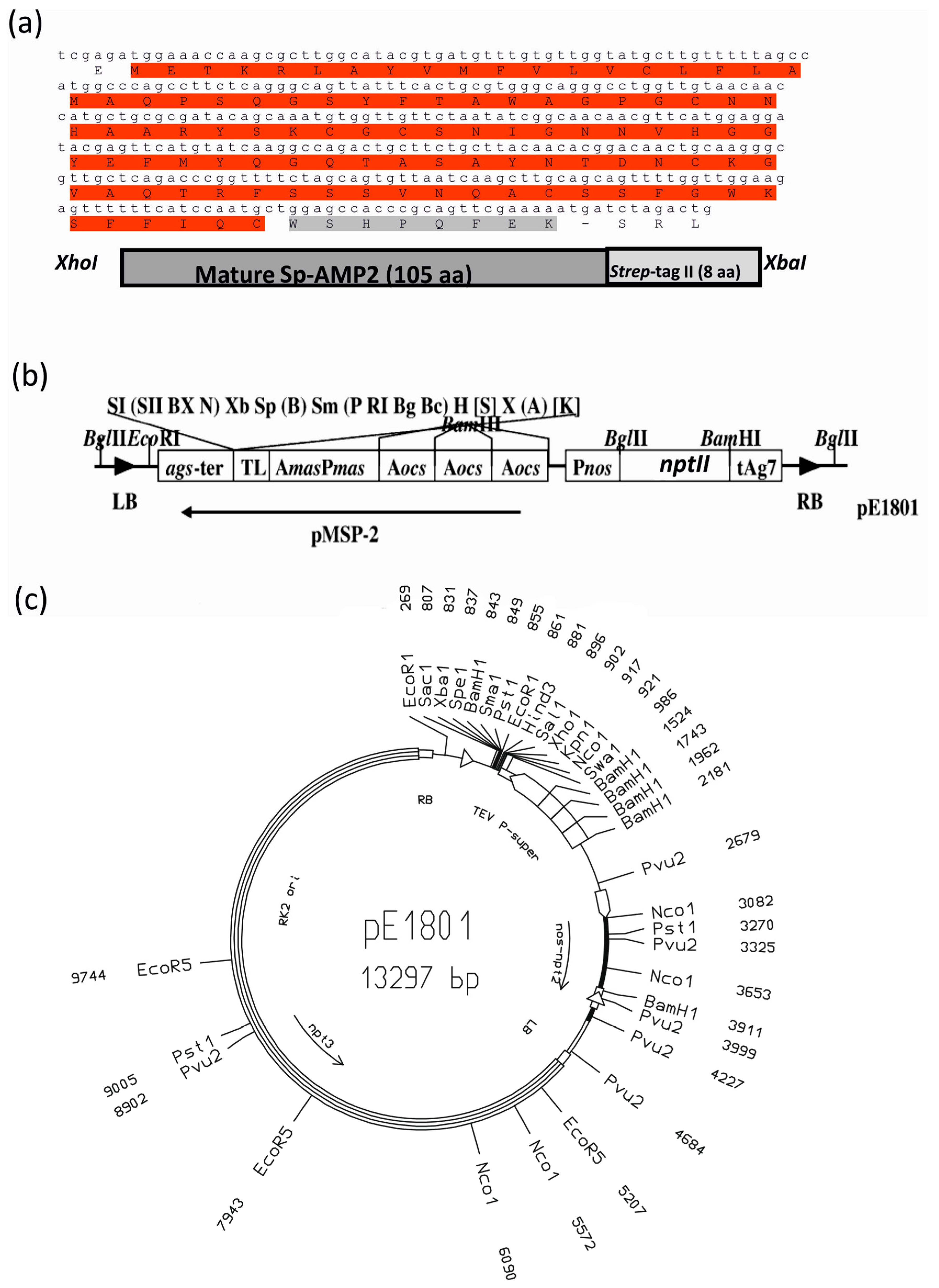
2.1. Agrobacterium Tumefaciens Vectors and Strains
2.2. Tobacco Transformation
2.3. Selection of Transgenic Plants
2.4. cDNA Synthesis by Reverse Transcription
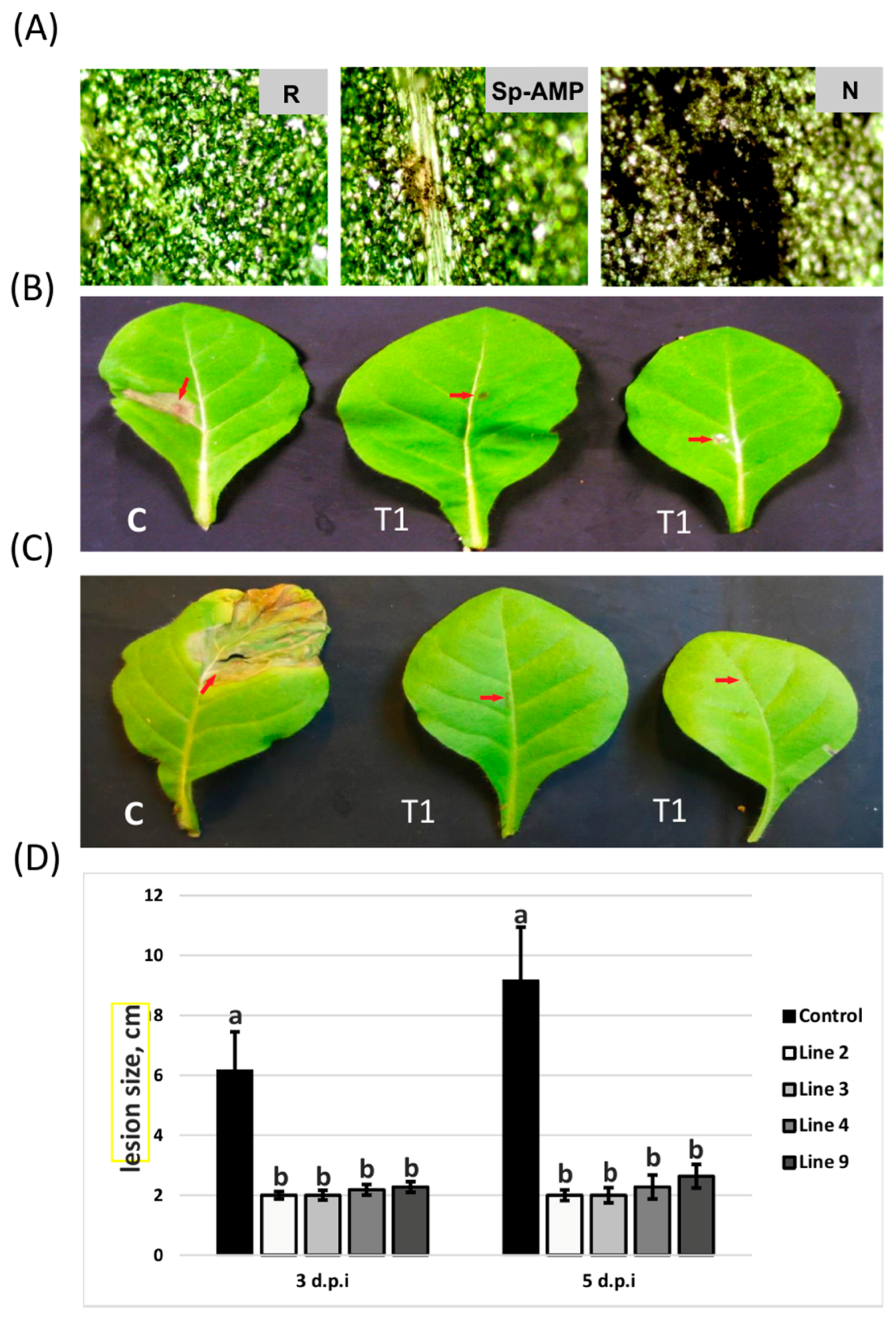
2.5. Pathogen Bioassays of Transgenic Plants
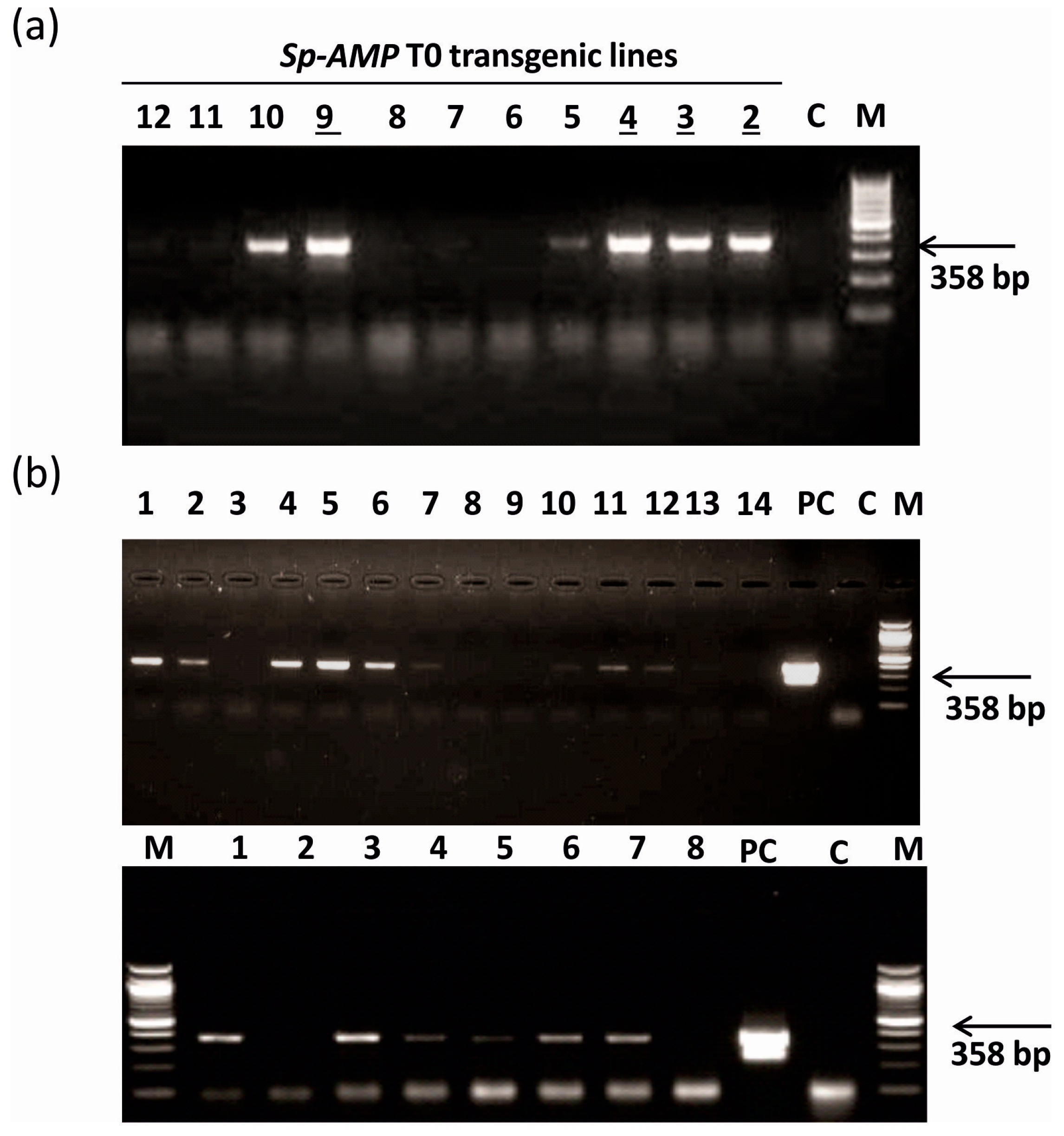
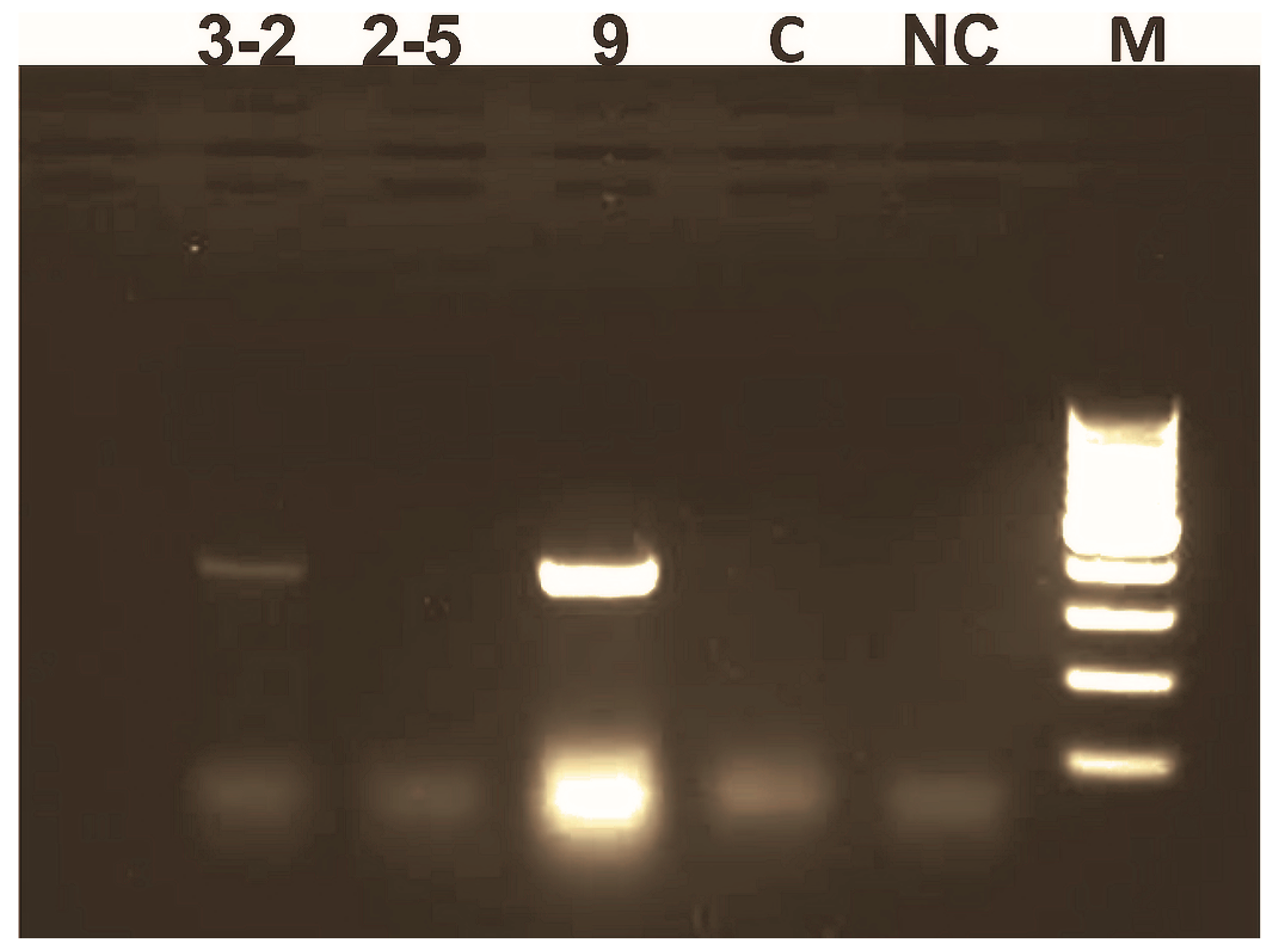
2.6. PCR Analysis of Transgenic Plants
2.7. Real-Time Quantitative PCR Analysis
2.8. Statistical Analyses
3. Results
3.1. Generation of Sp-AMP2 Transgenic Tobacco Plants
3.2. Sp-AMP2-Transformed Tobacco Plants Show Increased Tolerance to B. cinerea
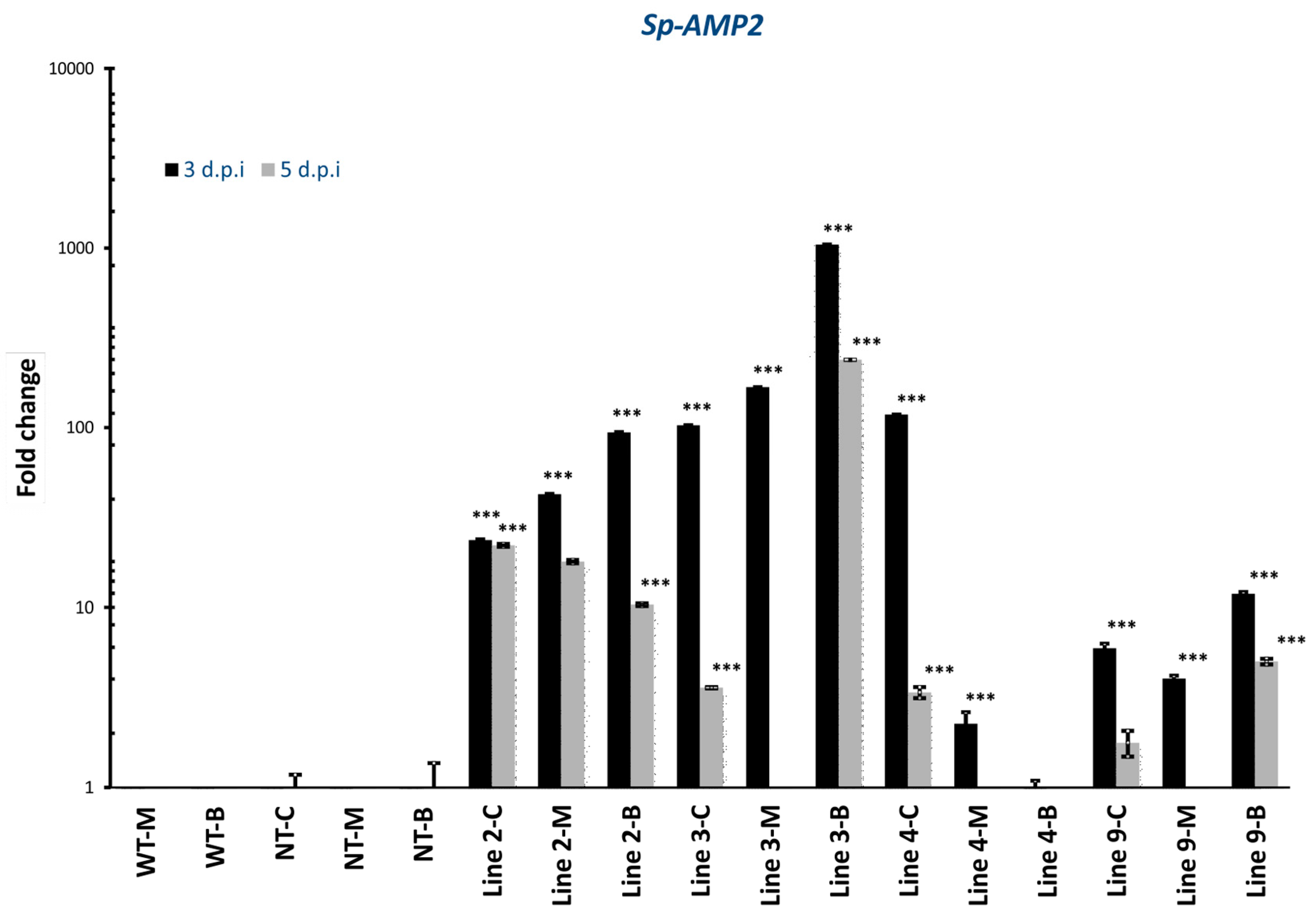
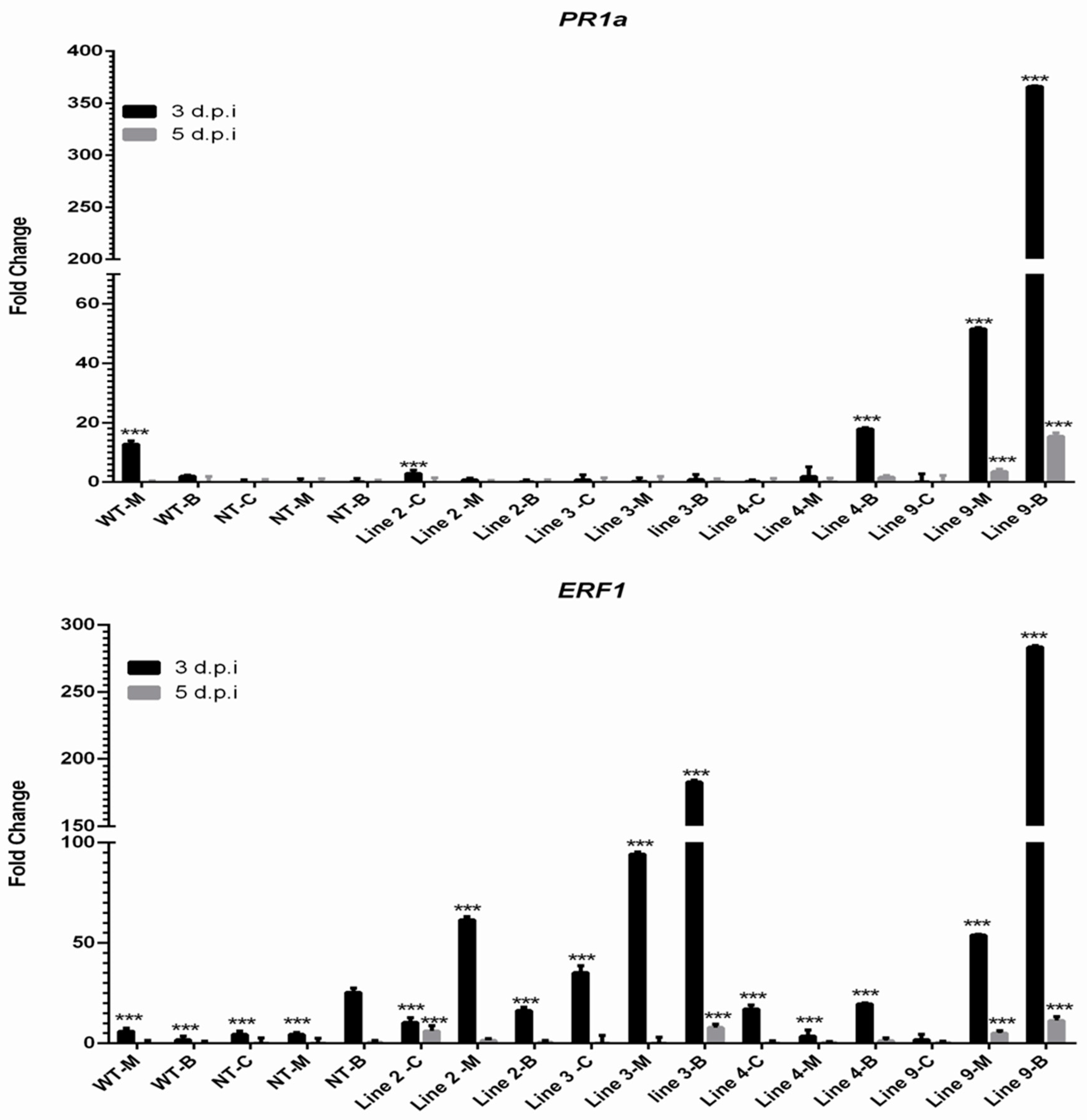
3.3. Expression Profiles of Sp-AMP2 Gene and SA/JA/ET Gene Markers in Transgenic Plants
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Cammue, B.P.A.; DeBolle, M.F.C.; Thevissen, K.; DeSamblanx, G.W.; Osborn, R.W. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 1997, 16, 297–323. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of pr-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Benko-Iseppon, A.M.; Galdino, S.L.; Calsa, T.; Kido, E.A.; Tossi, A.; Belarmino, L.C.; Crovella, S. Overview on plant antimicrobial peptides. Curr. Protein Pept. Sci. 2010, 11, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Padovan, L.; Scocchi, M.; Tossi, A. Structural aspects of plant antimicrobial peptides. Curr. Protein Pept. Sci. 2010, 11, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Ben Hamida, J.; Vergoten, G.; Fliss, I. Phytamp: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009, 37, D963–D968. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Jørgensen, H.J.L.; Lund, O.S.; Lyngkjær, M.F. Engineering pathogen resistance in crop plants: Current trends and future prospects. Annu. Rev. Phytopathol. 2010, 48, 269–291. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, E. Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett. 2007, 270, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, T.; Egorov, T. Plant antimicrobial peptides. In Plant Signaling Peptides; Irving, H.R., Gehring, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 107–133. [Google Scholar]
- Asiegbu, F.O.; Denekamp, M.; Daniel, G.; Johansson, M. Immune cytochemical-localization of pathogenesis-related proteins in roots of Norway spruce infected with Heterobasidion annosum. Eur. J. For. Pathol. 1995, 25, 169–178. [Google Scholar] [CrossRef]
- Davis, J.M.; Wu, H.G.; Cooke, J.E.K.; Reed, J.M.; Luce, K.S.; Michler, C.H. Pathogen challenge, salicylic acid, and jasmonic acid regulate expression of chitinase gene homologs in pine. Mol. Plant Microbe Interact. 2002, 15, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Asiegbu, F.O.; Daniel, G.; Johansson, M. Defense-related reactions of seedling roots of Norway spruce to infection by Heterobasidion annosum (Fr.) bref. Physiol. Mol. Plant Pathol. 1994, 45, 1–19. [Google Scholar] [CrossRef]
- Asiegbu, F.O.; Nahalkova, J.; Li, G.S. Pathogen-inducible cdnas from the interaction of the root rot fungus Heterobasidion annosum with scots pine (Pinus sylvestris L.). Plant Sci. 2005, 168, 365–372. [Google Scholar] [CrossRef]
- Mensen, R.; Hager, A.; Salzer, P. Elicitor-induced changes of wall-bound and secreted peroxidase activities in suspension-cultured spruce (Picea abies) cells are attenuated by auxins. Physiol. Plant. 1998, 102, 539–546. [Google Scholar] [CrossRef]
- Adomas, A.; Heller, G.; Li, G.; Olson, A.; Chu, T.M.; Osborne, J.; Craig, D.; van Zyl, L.; Wolfinger, R.; Sederoff, R.; et al. Transcript profiling of a conifer pathosystem: Response of Pinus sylvestris root tissues to pathogen (Heterobasidion annosum) invasion. Tree Physiol. 2007, 27, 1441–1458. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, V.; Kiyamova, R.; Cramer, R.; Krynytskyy, H.; Gout, I.; Filonenko, V.; Gout, R. Purification and molecular cloning of antimicrobial peptides from scots pine seedlings. Peptides 2009, 30, 2136–2143. [Google Scholar] [CrossRef] [PubMed]
- Fossdal, C.G.; Nagy, N.E.; Sharma, P.; Lonneborg, A. The putative gymnosperm plant defensin polypeptide (SPI1) accumulates after seed germination, is not readily released, and the SPI1 levels are reduced in pythium dimorphum-infected spruce roots. Plant Mol. Biol. 2003, 52, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Asiegbu, F.O.; Choi, W.; Li, G.; Nahalkova, J.; Dean, R.A. Isolation of a novel antimicrobial peptide gene (Sp-AMP) homologue from Pinus sylvestris (Scots pine) following infection with the root rot fungus Heterobasidion annosum. FEMS Microbiol. Lett. 2003, 228, 27–31. [Google Scholar] [CrossRef]
- Sooriyaarachchi, S.; Jaber, E.; Covarrubias, A.S.; Ubhayasekera, W.; Asiegbu, F.O.; Mowbray, S.L. Expression and beta-glucan binding properties of scots pine (Pinus sylvestris L.) antimicrobial protein (Sp-AMP). Plant Mol. Biol. 2011, 77, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Veluthakkal, R.; Dasgupta, M.G. Pathogenesis-related genes and proteins in forest tree species. Trees 2010, 24, 993–1006. [Google Scholar] [CrossRef]
- Manners, J.M. Primitive defence: The MiAMP1 antimicrobial peptide family. Plant Mol. Biol. Rep. 2009, 27, 237–242. [Google Scholar] [CrossRef]
- Kovalchuk, A.; Kerio, S.; Oghenekaro, A.O.; Jaber, E.; Raffaello, T.; Asiegbu, F.O. Antimicrobial defenses and resistance in forest trees: Challenges and perspectives in a genomic era. Annu. Rev. Phytopathol. 2013, 51, 221–244. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Cui, D.; Einstein, J.; Narasimhulu, S.; Vergara, C.E.; Gelvin, S.B. Strength and tissue-specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 1995, 7, 661–676. [Google Scholar] [CrossRef]
- An, G.; Ebert, P.; Mitra, A.; Ha, S. Binary vectors. In Plant Molecular Biology Manual; Gelvin, S., Schilperoort, R., Verma, D., Eds.; Springer: Dordrecht, The Netherlands, 1989; pp. 29–47. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general-method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Jaber, E.; Xiao, C.; Asiegbu, F.O. Comparative pathobiology of Heterobasidion annosum during challenge on Pinus sylvestris and arabidopsis roots: An analysis of defensin gene expression in two pathosystems. Planta 2014, 239, 717–733. [Google Scholar] [CrossRef] [PubMed]
- Quidde, T.; Buttner, P.; Tudzynski, P. Evidence for three different specific saponin-detoxifying activities in botrytis cinerea and cloning and functional analysis of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 1999, 105, 273–283. [Google Scholar] [CrossRef]
- Richards, E.; Reichardt, M.; Rogers, S. Preparation of genomic DNA from plant tissue. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.P.; Ryals, J.A. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Piqueras, R.; Sanchez-Serrano, J.J.; Solano, R. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Menda, N.; Tecle, I.Y.; Buels, R.M.; Strickler, S.; Fischer-York, T.; Pujar, A.; Leto, J.; Gosselin, J.; Mueller, L.A. The sol genomics network (solgenomics.Net): Growing tomatoes using perl. Nucleic Acids Res. 2011, 39, D1149–D1155. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Broekaert, W.F. Tissue-specific expression of plant defensin genes pdf2.1 and pdf2.2 in arabidopsis thaliana. Plant Physiol. Biochem. 1998, 36, 533–537. [Google Scholar] [CrossRef]
- Kazan, K.; Rusu, A.; Marcus, J.P.; Goulter, K.C.; Manners, J.M. Enhanced quantitative resistance to leptosphaeria maculans conferred by expression of a novel antimicrobial peptide in canola (Brassica napus L.). Mol. Breed. 2002, 10, 63–70. [Google Scholar] [CrossRef]
- Verma, S.S.; Yajima, W.R.; Rahman, M.H.; Shah, S.; Liu, J.J.; Ekramoddoullah, A.K.M.; Kav, N.N.V. A cysteine-rich antimicrobial peptide from pinus monticola (PmAMP1) confers resistance to multiple fungal pathogens in canola (Brassica napus). Plant Mol. Biol. 2012, 79, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Zamany, A.; Sniezko, R.A. Anti-microbial peptide (AMP): Nucleotide variation, gene expression, and host resistance in the white pine blister rust (WPBR) pathosystem. Planta 2013, 237, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Kazan, K.; Goulter, K.C.; Maclean, D.J.; Manners, J.M. The mode of action of the plant antimicrobial peptide MiAMP1 differs from that of its structural homologue, the yeast killer toxin WmKT. FEMS Microbiol. Lett. 2005, 243, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Elfstrand, M.; Fossdal, C.G.; Swedjemark, G.; Clapham, D.; Olsson, O.; Sitbon, F.; Sharma, P.; Lonneborg, A.; von Arnold, S. Identification of candidate genes for use in molecular breeding—A case study with the Norway spruce defensin-like gene, spi1. Silvae Genet. 2001, 50, 75–81. [Google Scholar]
- Walter, C.; Grace, L.J.; Donaldson, S.S.; Moody, J.; Gemmell, J.E.; van der Maas, S.; Kvaalen, H.; Lönneborg, A. An efficient biolistic® transformation protocol for picea abies embryogenic tissue and regeneration of transgenic plants. Can. J. For. Res. 1999, 29, 1539–1546. [Google Scholar] [CrossRef]
- Mentag, R.; Luckevich, M.; Morency, M.J.; Seguin, A. Bacterial disease resistance of transgenic hybrid poplar expressing the synthetic antimicrobial peptide d4e1. Tree Physiol. 2003, 23, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.P.; Goulter, K.C.; Green, J.L.; Harrison, S.J.; Manners, J.M. Purification, characterisation and cDNA cloning of an antimicrobial peptide from macadamia integrifolia. Eur. J. Biochem. 1997, 244, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Zamany, A.; Liu, J.J.; Ekramoddoullah, A.; Sniezko, R. Antifungal activity of a Pinus monticola antimicrobial peptide 1 (Pm-AMP1) and its accumulation in western white pine infected with Cronartium ribicola. Can. J. Microbiol. 2011, 57, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E.; Costa, L.M.; Gutierrez-Marcos, J. Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 2011, 62, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Nehra, N.S.; Becwar, M.R.; Rottmann, W.H.; Pearson, L.; Chowdhury, K.; Chang, S.J.; Wilde, H.D.; Kodrzycki, R.J.; Zhang, C.S.; Gause, K.C.; et al. Forest biotechnology: Innovative methods, emerging opportunities. In Vitro Cell. Dev. Biol.-Plant 2005, 41, 701–717. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaber, E.; Kovalchuk, A.; Raffaello, T.; Keriö, S.; Teeri, T.; Asiegbu, F.O. A Gene Encoding Scots Pine Antimicrobial Protein Sp-AMP2 (PR-19) Confers Increased Tolerance against Botrytis cinerea in Transgenic Tobacco. Forests 2018, 9, 10. https://doi.org/10.3390/f9010010
Jaber E, Kovalchuk A, Raffaello T, Keriö S, Teeri T, Asiegbu FO. A Gene Encoding Scots Pine Antimicrobial Protein Sp-AMP2 (PR-19) Confers Increased Tolerance against Botrytis cinerea in Transgenic Tobacco. Forests. 2018; 9(1):10. https://doi.org/10.3390/f9010010
Chicago/Turabian StyleJaber, Emad, Andriy Kovalchuk, Tommaso Raffaello, Susanna Keriö, Teemu Teeri, and Fred O. Asiegbu. 2018. "A Gene Encoding Scots Pine Antimicrobial Protein Sp-AMP2 (PR-19) Confers Increased Tolerance against Botrytis cinerea in Transgenic Tobacco" Forests 9, no. 1: 10. https://doi.org/10.3390/f9010010





