1. Introduction
Southamerican
Nothofagus spp. forests dominate temperate and sub-Antarctic regions of Chile and Argentina from 33° to 56° south latitude. In Chile, there are nine
Nothofagus species, including three evergreens and six deciduous species [
1,
2]. Second growth forests of roble (
Nothofagus obliqua (Mirb.) Oerst.), raulí (
N. alpina (Poepp. & Endl.) Oerst.), and coihue (
N. dombeyi (Mirb.) Oerst.), known locally as the “RORACO” forest type, are one of the most important native mixed forests of Chile. According to the most recent national forest inventory, there are about 1.4 million hectares of these forests in Chile accounting for 10.8% of the total native forest area of the country [
3]. In 2015 alone, the saw timber from RORACO forest type was approximately 45% of the native forest national timber production [
4]. Volume growth rates for these forests range between 6 and 10 m
3 ha
−1 year
−1 [
5] and are some of the highest rates for Southamerican temperate native forests. Hence, given its large geographical distribution, timber market value and attractive growth rates, these RORACO forests present high economic and social potential value within the Chilean forestry sector [
6].
RORACO forest type is highly variable over its large geographical area. This variability is in the form of flora and faunal diversity, varying management regimes, anthropogenic disturbances, and site productivity levels [
7]. For example, the three
Nothofagus species typically have contrasting altitude ranges:
N. obliqua is commonly found between 100 and 600 m a.s.l.,
N. alpina between 600 and 900 m a.s.l., and
N. dombeyi is frequently found over 900 m a.s.l, and it is possible to find different ecotones as single or mixed composition forests [
8]. Echeverria and Lara [
9] found that the main environmental factors affecting RORACO growth rates are: longitude, climatic variables (e.g., mean annual rainfall, summer humidity index, frost-free period), and soil quality (e.g., texture), which alone accounted for 70.4% of the total observed variance. Defining appropriate growth zones can help in managing for this variability, as there are a few studies that have done this for the RORACO forest type [
9,
10,
11]. Overall, these studies found that within a given growth zone, RORACO forests are characterized by similar growth patterns, silvicultural treatment responses, and associated species. However, due to this large stand variability, predictive and improved decision-making tools and models for these forests are scarce and often of poor quality.
Forest inventories provide with critical information of current stand conditions, but growth and yield (G & Y) models provide improved information on future conditions and help modeling forest dynamics and management regimes [
12]. For the species that conform to RORACO forest, some specific biometric studies have been reported; however, often these are applicable to restricted geographical areas and for specific environmental conditions [
6,
9,
10,
11,
13,
14,
15].
G & Y models are useful tools for inventory updates and decision making regarding short and long-term silvicultural management targets [
16]. G & Y models have been developed mainly for plantations or even aged and single species forests, as the complexity for mixed or uneven-aged forests increases considerably, there are not many of such models. Blanco et al. [
17] indicated that G & Y models for mixed-forest have been more numerous in North America and Central Europe; the four models more popular are: FORESCAST, SILVA, FORMIX and FORMIND, where the latter has been previously used for Chilean temperate rain forests [
17]. Some of the challenges in developing G & Y models for mixed-forest include: growth rates that differ among species, stands that include shade tolerant and intolerant species, and requirement for selective harvests with a continuous forest cover. These, and other factors, increase uncertainty and complexity of these systems and lower their prediction accuracy [
18].
Several G & Y modelling approaches have been developed based on: initial stand conditions, species composition, and forest management objectives [
18]. These approaches range from models that provide aggregated stand variables to resolutions as high as the individual tree [
18]. Stand-level are the most common G & Y models, and these can be expanded to produce a stand table via the generation of a diameter distribution with several size classes [
19]. Stand-level models often provide accurate estimates and projections of the overall stand variables [
18,
20], but at a low level of resolution (i.e., stand). Individual-tree models offer high flexibility to describe and project any stand structure and allow for the incorporation of reasonable process-based modules, such as thinning, differentiation of growth patterns and, in the case of mixed forest, individual responses for each species to competition and mortality. There are two types of individual-tree models: distance-dependent or distance-independent model, which differ in the inclusion of the physical coordinates of each tree within a plot. Both types have been widely used for different forest types, including plantations, even-aged mixed hardwood stands, uneven-aged mixed species, tropical forests, boreal forests, and temperate forests [
14,
21,
22,
23,
24,
25,
26,
27]. Nevertheless, individual-tree models for projecting growth and yield are data intensive, but they can be used as feedback calibration with stand-level models [
12,
18,
28]. Individual-tree models present good results for short-term projections, with a similar level of accuracy respect to whole-stand models [
28,
29]. However, these models often have poorer levels of accuracy for long-term projections, where whole-stand models are more reliable. [
24]. Salas et al. [
14] mentioned that both distance-dependent and distance-independent models do not have significant differences in growth prediction, even in natural forests; furthermore, spatial dependency is often more complex to model. An individual-tree G & Y model consists of three components [
18]: (1) an individual-tree diameter breast height (DBH) or basal area growth equation; (2) an individual-tree height growth equation or, alternatively, a height-diameter relationship; and (3) a mortality or survival component. Basal area or DBH growth can be modeled using a function of many tree and stand attributes [
18,
27,
30]. For mixed forests and large forest areas these growth equations are calibrated to include different species and/or geographical zones [
25]. Mortality/survival equations often use logistic models that also consider individual-tree attributes together with a measure of tree competition and stand attributes [
18,
22,
26,
27].
The majority of the G & Y models are obtained by fitting multiple linear regressions with a suite of potential predictors in their original units or transformed. However, these predictors are often characterized by high levels of multicollinearity, making the identification of the relevant predictors more difficult [
31]. Hence, incorporating variable selection procedures in G & Y model development could help in the identification of relevant predictors, and therefore, improve both the statistical validity of the final model and its usefulness in forest management decisions.
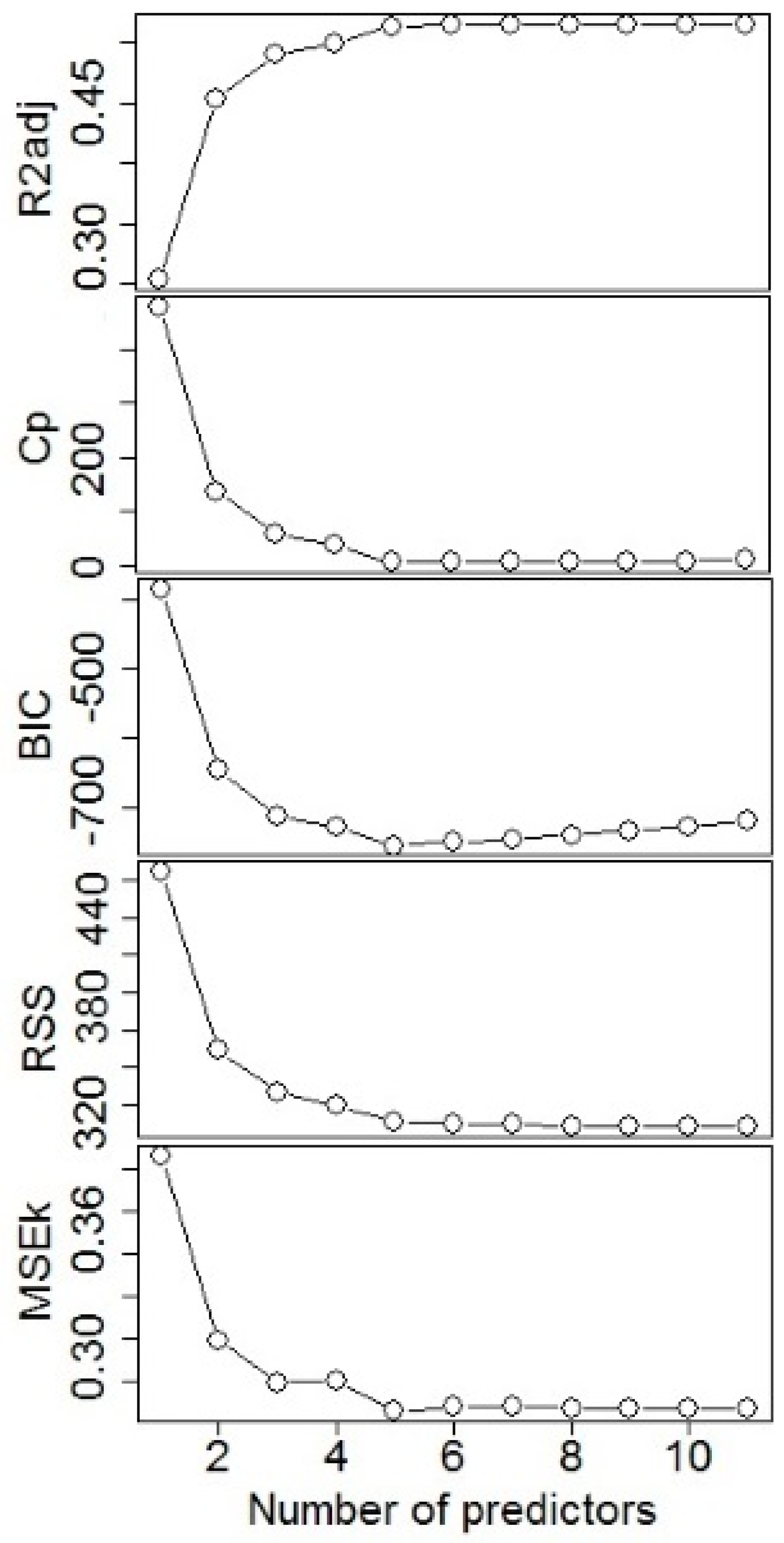
There are several statistical methods for selecting the best predictors including: best subset, forward, backward, forward stepwise, backward stepwise, and hybrid selection procedures [
32]. These selection methods have the same logic, which is to fit a linear model with the selected subset of predictors [
33]. Alternatively, a shrinkage or regularization method has been used, where a linear model is fitted with all predictors simultaneously incorporating a constraint that shrinks model coefficients towards zero. The advantage of these procedures is the reduction of the variance of each coefficient estimate [
32,
33,
34,
35]. Here, the two best-known procedures are: ridge regression (RR) and least absolute shrinkage and selection operator (LASSO) [
33,
35]. Both procedures are similar to least squares, except for the incorporating of a shrinkage penalty associated with a λ tuning parameter. The advantage of the LASSO is that it makes some coefficient estimates exactly zero, particularly when the tuning parameter λ is large; thus, LASSO at the same time performs shrinkage and variable selection [
32,
33,
35]. One limitation of LASSO is that under non-Bayesian statistics, due to the penalized estimation, it does not provide p-values, confidence intervals or standard error of the regression coefficients [
34,
36,
37].
The majority of G & Y models are created to respond to scientific problems, rather than to support management decision; furthermore, many of the G & Y models are limited to their measurement conditions [
17]. On the other hand, temperate ecosystems are the biomes with more available G & Y models for mixed-forest (55%); however, in relation to other geographical zones, Southamerican temperate forests have the lowest availability of models [
17]. Given this lack of tree and forest growth information and the importance of the RORACO forest type in Chile, there is a need for improved empirical growth models than can account for the variability and diverse geographic distribution of this resource (>14,000 km
2, including four growth zones). Thus, the aim of this study is to develop an individual-tree growth model to estimate annual increment in DBH for second growth mixed forest stands of
N. alpina,
N. obliqua, and
N. dombeyi, including a set of tree- and stand-level predictors to explain diameter growth. The specific objectives of this study are: (1) to compare different multiple linear fitting procedures to obtain the best individual-tree growth model; (2) to generalize diameter growth models by incorporating growth zone and species; and (3) to evaluate the final diameter growth model using an independent validation dataset. Accordingly, to meet these objectives, advanced statistical model predictor selection methods are used in currently available forest inventory and plot data that spans RORACO’s ecologically and geographically diverse area.
4. Discussion
Diameter growth models have been recognized as useful for projecting individual-tree growth [
18,
19,
40]. In this study, a DBH growth distance-independent model was developed for mixed
Nothofagus second growth forest in southern Chile. The response variable for this study was annual average DBH growth obtained over a period of two years that originated from increment cores and tree sections. Several important aspects are associated with the use of this response variable. First, a short time period of two years was chosen to improve the association of observed tree growth with its present tree and stand conditions, a relationship that weakens as the period gets longer. This is often critical for those years with ecological disturbances (e.g., climatic, diseases, catastrophic event); however, Andreassen and Tomter [
24] found no effects on the prediction accuracy with different growth period lengths. Second, no evidence of wood compression was found in increment cores when compared to sections. Finally, individual basal area growth was discarded as a response as it presented poorer goodness-of-fit statistics than DBH growth (data not shown).
Individual-tree growth models that include predictors of competition, productivity, tree- and stand-level have been used in many reported studies [
21,
22,
23,
24,
25,
26,
27,
49]. Hence, there are potentially many predictors that might describe the same biological factor, which will suffer from multicollinearity producing inconsistences and unexpected results in the fitted models [
31]. Thus, it is necessary to select an adequate subset of these predictors using some variable selection procedure. In this study, the fitting database contained, as expected, pairs of predictors with high correlation, such as BA-BAN, Hd-RS, BA-SDI, BAL-BALn, Hd-QD, A-Ad, RS-QD, and DBH-H, with correlation values greater than 0.83, and this required the implementation of appropriate variable selection procedures such as CV and LASSO regression that deal with multicollinearity.
The predictors selected by CV and LASSO regression corresponded to competition, tree- and stand-level variables, but they did not included predictors associated with productivity. In CV regression DBH, A, SS, SDI, and BALn were the selected predictors, while in LASSO regression these were the same with the addition of Ad and BALr. The tree-level variables selected corresponded to DBH and A, both mostly measures of tree size; where, according to the estimated parameters, growth rates increase with larger diameters; however, age had a negative slope, indicating that at a fixed DBH value, older trees present lower growth rates. This agrees with a similar result reported by Cubillos [
50] in
N. alpina.
In our study, competition variables had a high relevance in explaining individual-tree growth, as found in other
Nothofagus studies [
50,
51,
52,
53,
54]. Interestingly, the predictor BALn (basal area of larger trees only for
Nothofagus) was selected instead of BAL (that includes all species). The selection of BALn indicates that light, water and nutrients competition is related to individuals of the same cohort, in this case, the first strata of
Nothofagus for one side and the companion species for the other. This ‘independence’ between these groups is mostly due to the individual strategies of each cohort, where some are shade intolerant (
Nothofagus) and others shade tolerant (companion species) eliminating, or reducing, competition for light. Competition for water and nutrients is also expected to be limited as it is related to roots depth, where
Nothofagus are older than the companion species using greater soil depth [
2,
55]. However, the fine root biomass for
N. dombeyi is concentrated in the first 30 cm of depth sharing the same space that companion species, thus the no nutrients competition between this two cohorts are related to the effect of ectomycorrhizal (
Nothofagus), arbuscular mycorrhizae (companion species) and fertility of this volcanic soils [
55,
56]. This null or limited competition between cohorts agrees with findings from previous reports that support the theory of additive effects in growth for
Nothofagus and companion species [
55,
57,
58], that translates into independent behavior (and therefore models) between these two cohorts. Lastly, productivity predictors, such as SI, were not selected for modelling DBH growth in these forests, which is probably due to several factors, such as uncertainty on the quality of dominant height-site models, and measurement errors on dominant height and age, among others.
The average DBH growth rates for combinations of species and zone varied considerably, with important differences between species and zone; however, extreme cases where noted for the group combinations of
N. alpine—zone 1, and
N. dombeyi—zone 3, with an 88% larger growth rate for the latter (2.15 mm year
−1 against 4.05 mm year
−1). Other studies also have found contrasting differences in growth between
Nothofagus species and growth zones [
5,
9,
10,
59,
60,
61,
62,
63,
64]. The incorporation of species and/or zone factors to improve growth models has been previously reported [
24,
25]. In the present study, the combined specie-zone factor (SpZone, with 11 levels), explained more variability than the individual two factors (with seven levels for both species and zone), indicating that a greater disaggregation of the data is required to model growth rate accurately. Interestingly, the LASSO + SpZone model selected as predictors for the combined factor SpZone the extreme groups (
Table 6), by combining the central five levels into a single group class.
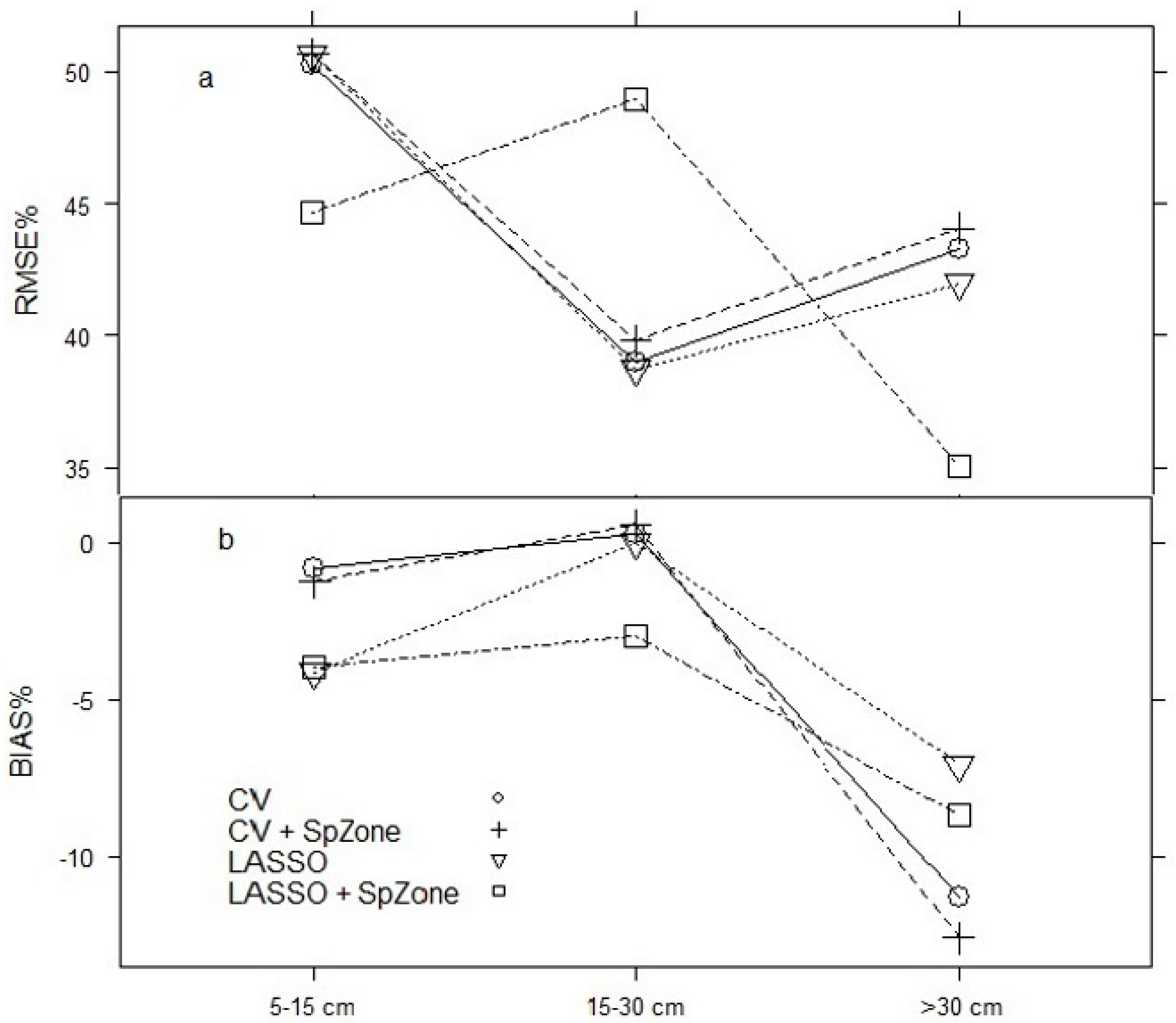
The fitting of the four candidate models to predict AIDBH resulted in overall good performance, with R
2emp ranging from 0.54 to 0.57, RMSE% of ~44%, and a BIAS% smaller than 3%. Similar goodness-of-fit statistics have been reported in other species for AIDBH models with R
2emp ranging from 0.26 to 0.68 [
24,
26,
51,
65]. AIDBH projections for 6 and 12 years resulted in R
2emp ranges of 0.23–0.28 and 0.15–0.24, respectively. This drop in performance is likely to have been affected by the quality of the projection database, that included, for example, 6 years with 20% of the trees with null or negative DBH increments, probably due to field measurement errors and low growth rates for these species. In contrast, for both
Nothofagus and companion species, DBH projections for 6 and 12 years resulted in overall R
2emp values greater than 0.97. Small diameter trees (DBH < 15 cm) presented lower correlations, with values of 0.88 and 0.58 for projections of 6 and 12 years, respectively. In addition, as expected, goodness-of-fit statistics were better for the
Nothofagus cohort than the companion species cohort.
The final selected model in this study is CV + SpZone (Equation (10)). The LASSO model was discarded, even though it was highly competitive, due to lower prediction performance, and it also included some predictors with high multicollinearity (VIF > 5). LASSO + SpZone had the worst performance with issues for prediction and projection. The CV model presented better goodness-of-fit statistics than the CV + SpZone model; however, the combined factor SpZone was highly significant (
p < 0.001). Hence, the inclusion of SpZone is strongly justified given the wide geographical range of this population and its differential diameter growth responses. This large stand variability has also been detected before, leading to many authors defining growth zones for this resource [
9,
10,
11]. Alternatively, with no information of growth zone, it is possible to use the simple CV model that will still provide reliable predictions of AIDBH.
The simplified final selected model corroborates well known forest ecological processes and basic biological consistency in the parameter estimates (
Table 2). For example, BALn and SS are competition variables where a higher value indicates a high competition for a given tree, and their model parameters both have a negative sign; hence, with higher competition there are lower growth rates. In the case of the inverse of BALr, the estimated coefficient value is positive, thus a high competition produces a lower growth rate, and vice versa. The most important predictor, DBH, has a strong effect on growth, where larger trees have a greater diameter increment due to its positive sign, which is expected as this effect is controlled indirectly by age. Hence, at the same age, larger trees tend to have greater growth rates. Finally, the parameter associated with age was negative, thus for a fixed DBH value, a young tree is capable of higher growth rates than an older tree.
The advantages of CV procedure as a selection method is the possibility of obtaining an independent estimation of the mean square error (MSE
K) when compared to other stepwise methods. LASSO is an alternative procedure that regularizes the coefficient estimates, forcing some of them to be exactly equal to zero, and hence, performing variable selection. Both procedures, CV and LASSO, have been used broadly due their advantages to identify those variables that help to better predict a given response [
32]. Similarly, the importance of incorporating dummy variables (or factors) in a linear model helps to expand model specificity, where additional factors can modify intercepts or slopes from the original regression model to make obtain more generic, and therefore robust, models that apply for wider conditions.
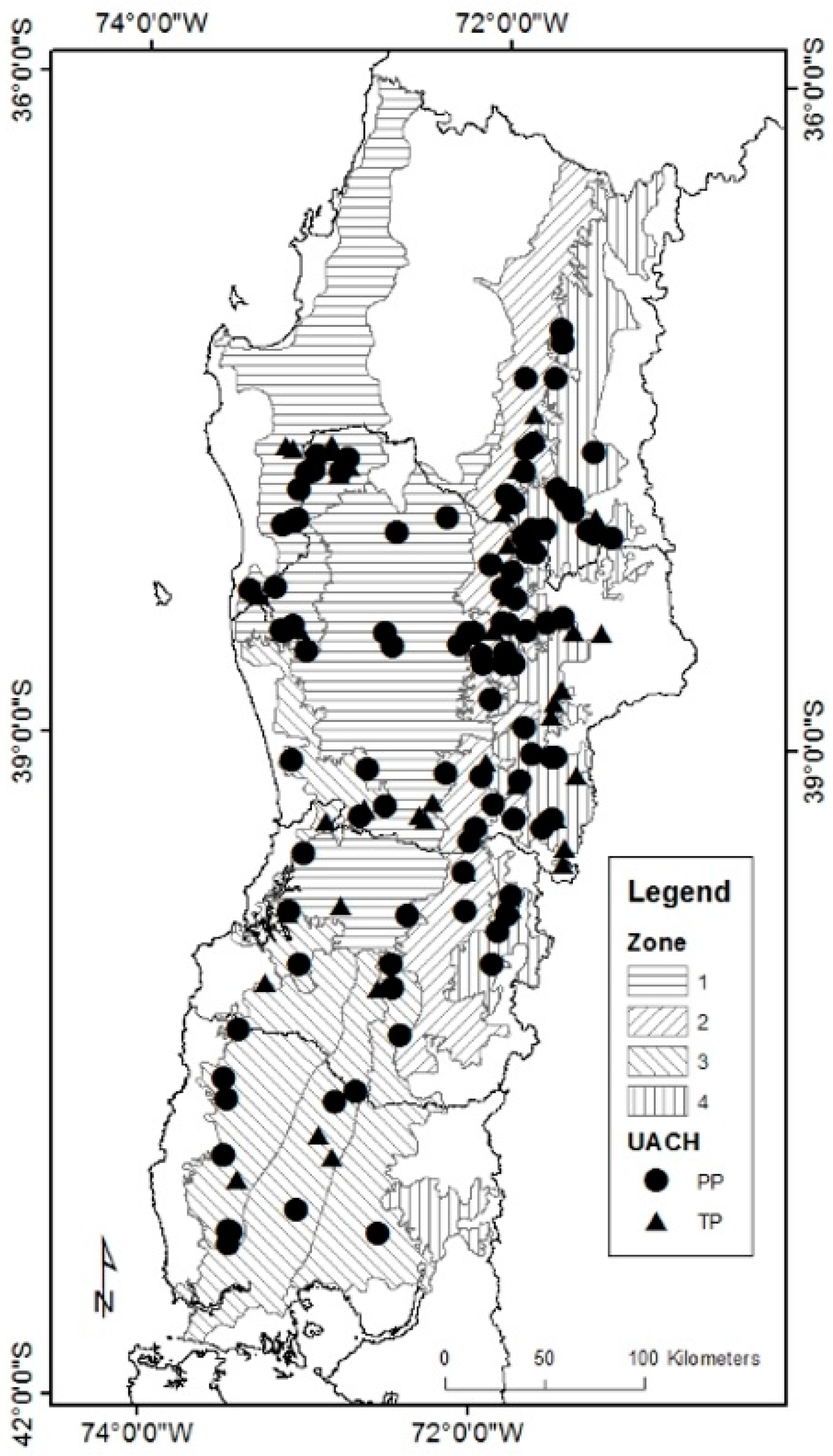
The final reported model should be used within the temporal and spatial frame of inference for RORACO second growth forests located between 37° and 41.5° south latitude, with breast height ages ranging from 20 to 80 years, and with a
Nothofagus basal area greater than 60% of the total basal area, due to the characteristics of the database used in this study (
Table 1). Similarly, it is recommended that this model is used for projections no longer than 12 years. For this projection period, the temporal frame of inference for this model is well suited for existing forest management plans in Chile that typically consider durations of 10 years, among a range of silvicultural interventions.
Future research is warranted to better improve the performance of models of individual-tree growth for this population. Some of these extensions could include: (1) producing models that do not require age, as this predictor is often difficult to obtain from typical forest inventories for natural stands; (2) including extra validation data from a wider geographic range and with additional forest conditions; (3) developing an individual-tree growth model for companion species; (4) implementing a fully individual-tree model routine that includes a mortality and a height growth module; and (5) developing a compatible system based on both individual-tree and stand-level models.