Estimating Forest Characteristics for Longleaf Pine Restoration Using Normalized Remotely Sensed Imagery in Florida USA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Overview
2.3. Sample Design
2.4. Field Data
2.5. Image Normalization
2.6. Spectral and Texture Metrics
2.7. Model Development, Comparisons, and Raster Surface Creation
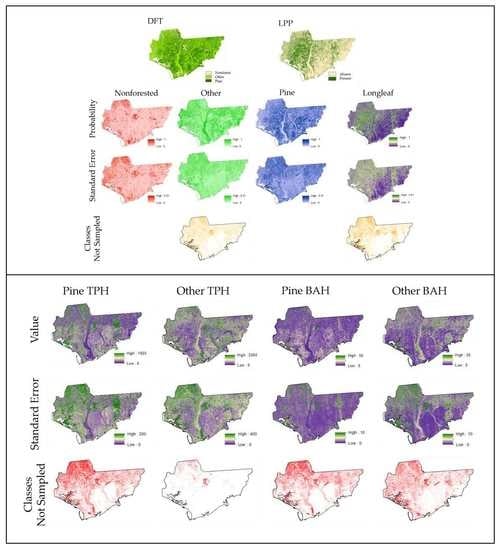
3. Results
3.1. Field Data
3.2. Image Normalization
3.3. Sample Distribution
3.4. Model Development, Comparisons, and Raster Surface Creation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

| Class | FIA Species Codes |
|---|---|
| Pine | 115, 128, 131, 107, 110, 111, 121 (Longleaf pine) |
| Other | 819, 835, 838, 842, 806, 824, 840, 841, 68, 221, 222, 802, 812, 813, 820, 825, 827, 831, 837, 822, 833, 834, 461, 462, 491, 521, 531, 544, 555, 591, 611, 621, 652, 653, 682, 691, 692, 693, 694, 711, 721, 762, 858, 922, 931, 972, 975, 993, 999, 316, 373, 391, 311, 317, 323, 345, 356, 367, 381, 421, 471, 500, 502, 541, 548, 551, 552, 581, 660, 662, 681, 701, 722, 731, 760, 764, 766, 901, 912, 953, 971, 994, 402, 403, 404, 409, 401, 410 |
| Source | Season | Path\Row\Tile | Acquisition Date |
|---|---|---|---|
| Landsat 8 | LDS | 18\39+ | 10/11/2016 |
| Landsat 8 | LDS | 19\39 | 10/16/2015 |
| Landsat 8 | LGS | 18\39+ | 5/7/2017 |
| Landsat 8 | LGS | 19\39 | 4/9/2016 |
| Landsat 8 | LWS | 18\39+ | 1/18/2018 |
| Landsat 8 | LWS | 19\39 | 12/21/2016 |
| Sentinel 2 | LDS | 16RFV+ | 10/12/2018 |
| Sentinel 2 | LDS | 16RFU+ | 10/12/2018 |
| Sentinel 2 | LDS | 16RFT+ | 10/12/2018 |
| Sentinel 2 | LDS | 17RGU | 10/14/2018 |
| Sentinel 2 | LDS | 17RKP | 10/14/2018 |
| Sentinel 2 | LGS | 16RFV | 3/16/2017 |
| Sentinel 2 | LGS | 16RFU | 3/16/2017 |
| Sentinel 2 | LGS | 16RFT+ | 5/2/2017 |
| Sentinel 2 | LGS | 17RGU+ | 5/2/2017 |
| Sentinel 2 | LGS | 17RKP+ | 5/2/2017 |
| Sentinel 2 | LWS | 16RFV+ | 1/30/2018 |
| Sentinel 2 | LWS | 16RFU+ | 1/30/2018 |
| Sentinel 2 | LWS | 16RFT | 2/24/2017 |
| Sentinel 2 | LWS | 17RGU | 1/12/2017 |
| Sentinel 2 | LWS | 17RKP | 1/12/2017 |
| Aerial Imagery | LDS | 7001 | 10/26/2017 |
| Aerial Imagery | LDS | 7002 | 10/26/2017 |
| Aerial Imagery | LDS | 7003 | 10/26/2017 |
| Aerial Imagery | LDS | 7004 | 10/26/2017 |
| Aerial Imagery | LDS | 8001 | 11/10/2017 |
| Aerial Imagery | LDS | 8002 | 11/18/2017 |
| Aerial Imagery | LDS | 8003 | 11/18/2017 |
| Aerial Imagery | LDS | 8004 | 11/18/2017 |
| Aerial Imagery | LDS | 8005 | 11/5/2017 |
| Aerial Imagery | LDS | 8006 | 11/5/2017 |
| Aerial Imagery | LDS | 8007 | 11/5/2017 |
| Aerial Imagery | LDS | 8008 | 11/5/2017 |
| Aerial Imagery | LDS | 8009 | 11/5/2017 |
| Aerial Imagery | LDS | 8010 | 11/1/2017 |
| Aerial Imagery | LDS | 8011 | 11/1/2017 |
| Aerial Imagery | LDS | 8012 | 11/1/2017 |
| Aerial Imagery | LDS | 8013 | 11/1/2017 |
| Aerial Imagery | LDS | 8014 | 10/26/2017 |
| Aerial Imagery | LDS | 9017 | 10/29/2017 |
| Aerial Imagery | LDS | 9018 | 10/24/2017 |
| Aerial Imagery | LDS | 9019 | 10/24/2017 |
| Aerial Imagery | LDS | 9020 | 10/24/2017 |
| Aerial Imagery | LDS | 9021 | 10/24/2017 |
| Aerial Imagery | LDS | 9022 | 10/24/2017 |
| Aerial Imagery | LDS | 9023 | 10/24/2017 |
| Aerial Imagery | LDS | 9024 | 10/24/2017 |
| Aerial Imagery | LDS | 9025 | 10/24/2017 |
| Aerial Imagery | LDS | 9026 | 10/24/2017 |
| Aerial Imagery | LDS | 9027 | 10/24/2017 |
References
- Noss, R.; LaRoe, E.; Scott, J. Endangered Ecosystems of the United States: A Preliminary Assessment of Loss and Degradation; Biological Report 28; National Biological Service: Washington, DC, USA, 1995; Available online: https://sciences.ucf.edu/biology/king/wp-content/uploads/sites/106/2011/08/Noss-et-al-1995.pdf (accessed on 19 July 2019).
- Oswalt, C.; Cooper, J.; Brockway, D.; Brooks, H.; Walker, J.; Connor, K.; Oswalt, S.; Conner, R. History and Current Condition of Longleaf Pine in the Southern United States; USDA Forest Service General Technical Report SRS-166; United States Forest Service: Ashville, NC, USA, 2012. Available online: http://www.srs.fs.usda.gov/pubs/42259 (accessed on 6 February 2019).
- Regional Working Group for America’s Longleaf. Range-Wide Conservation Plan for Longleaf. 2009. Available online: http://www.americaslongleaf.org/media/86/conservation_plan.pdf (accessed on 6 February 2019).
- U.S. Forest Service Forest Inventory and Analysis Program: We Are the Nation’s Forest Census. Available online: https://www.fia.fs.fed.us/ (accessed on 6 February 2019).
- Hogland, J.; Anderson, N.; St. Peter, J.; Drake, J.; Medley, P. Mapping forest characteristics at fine resolution across large landscapes of the southeastern United States using NAIP imagery and FIA field plot data. ISPRS Int. J. Geo-Inf. 2018, 7, 140. Available online: https://www.mdpi.com/2220-9964/7/4/140/htm (accessed on 6 February 2019). [CrossRef] [Green Version]
- Hogland, J.; Anderson, N.; Affleck, D.L.R.; St. Peter, J. Using Forest Inventory Data with Landsat 8 imagery to Map Longleaf Pine Forest Characteristics in Georgia, USA. Remote Sens. 2019, 11, 1803. [Google Scholar] [CrossRef] [Green Version]
- Gibert, K.; Horsburgh, J.S.; Athanasiadis, I.N.; Holmes, G. Environmental Data Science. Environ. Model. Softw. 2018, 106, 4–12. [Google Scholar] [CrossRef]
- Homer, C.; Dewitz, J.; Yang, L.; Jin, S.; Danielson, P.; Xian, G.; Coulston, J.; Herold, N.; Wickham, J.; Megown, K. Completion of the 2011 National Land Cover Database for the conterminous United States-Representing a decade of land cover change information. Photogr. Eng. Remote Sens. 2015, 81, 345–354. [Google Scholar]
- LANDFIRE. Existing Vegetation Type Layer, LANDFIRE 1.1.0, U.S. Department of the Interior, Geological Survey. 2008. Available online: http://landfire.cr.usgs.gov/viewer/ (accessed on 28 October 2010).
- Brunner, R.J.; Kim, E. Teaching Data Science. Procedia Comput. Sci. 2016, 80, 1947–1956. [Google Scholar] [CrossRef] [Green Version]
- Lokers, R.; Knapen, R.; Janssen, S.; van Randen, Y.; Jansen, J. Analysis of Big Data technologies for use in agro-environmental science. Environ. Model. Softw. 2016, 84, 494–504. [Google Scholar] [CrossRef] [Green Version]
- The Longleaf Alliance. About ARSA. Available online: https://www.longleafalliance.org/arsa/about-arsa (accessed on 23 October 2019).
- Quantum Spatial. About Use. Available online: https://www.quantumspatial.com/about-us (accessed on 23 October 2019).
- Earth Observing System [EOS]. Sentinel-2. Available online: https://eos.com/sentinel-2/ (accessed on 23 October 2019).
- United States Geological Survey [USGS]. Landsat 8. Available online: https://www.usgs.gov/land-resources/nli/landsat/landsat-8?qt-science_support_page_related_con=0#qt-science_support_page_related_con (accessed on 23 October 2019).
- USGS. Landsat 8 Surface Reflectance Code (LASRC) Product Guide. General Technical Report LSDS-1368 Version 2.0. 2019. Available online: https://prd-wret.s3-us-west-2.amazonaws.com/assets/palladium/production/atoms/files/LSDS-1368_L8_Surface_Reflectance_Code_LASRC_Product_Guide-v2.0.pdf (accessed on 19 July 2019).
- Florida Natural Areas Inventory [FNAI]. About Us. Available online: https://www.fnai.org/about.cfm (accessed on 23 October 2019).
- ESA Sentinel Online. Copernicus Open Access Hub. Available online: https://scihub.copernicus.eu/dhus/#/home (accessed on 23 October 2019).
- USGS. EarthExplorer—Home. Available online: https://earthexplorer.usgs.gov/ (accessed on 23 October 2019).
- Hogland, J.; Affleck, D.L.R. Mitigating the Impact of Field and image Registration Errors through Spatial Aggregation. Remote Sens. 2019, 11, 222. [Google Scholar] [CrossRef] [Green Version]
- Souza, C. Accord.Net Framework. Available online: http://accord-framework.net/ (accessed on 27 September 2013).
- Hogland, J.; Anderson, N. Function Modeling Improves the Efficiency of Spatial Modeling Using Big Data from Remote Sensing. Big Data Cogn. Comput. 2017, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- Hogland, J. Creating Spatial Probability Distributions for Longleaf Pine Ecosystems Across East Mississippi, Alabama, The Panhandle of Florida, and West Georgia, Thesis. 2005. Available online: https://etd.auburn.edu/bitstream/handle/10415/603/HOGLAND_JOHN_19.pdf?sequence=1&isAllowed=y (accessed on 20 December 2017).
- Elvidge, C.D.; Yuan, D.; Weerackoon, R.D.; Lunneta, R.S. Relative radiometric normalization of Landsat Multispectral Scanner (MSS) data using an automatic scattergram-controlled regression. Photogramm. Eng. Remote Sens. 1995, 61, 1255–1260. [Google Scholar]
- Wood, S.N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. 2011, 73, 3–36. [Google Scholar] [CrossRef] [Green Version]
- Wood, S.N.; Augustin, N.H. GAMs with integrated model selection using penalized regression splines and applications to environmental modeling. Ecol. Model. 2002, 157, 157–177. [Google Scholar] [CrossRef] [Green Version]
- Akaike, H. Information theory and an extension of the maximum likelihood principle. In Proceedings of the 2nd International Symposium on Information Theory, Tsahkadsor, Armenia, 2–8 September 1971; Petrov, B.N., Csaki, F., Eds.; Akadémiai Kiadó: Budapest, Hungary, 1973; pp. 267–281. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Moran, P. Notes on Continuous Stochastic Phenomena. Biometrika 1950, 37, 17–23. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 28 April 2018).
- Cressie, N.A.C. Statistics for Spatial Data, Revised ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1993; p. 928. [Google Scholar]
- Tille, Y.; Wilhelm, M. Probability Sampling Designs: Principles for Choice of Design and Balancing. Stat. Sci. 2017, 32, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Gregoire, T.; Valentine, H. Sampling Strategies for Natural Resources and the Environment; Chapman & Hall: Boca Raton, FL, USA; London, UK; New York, NY, USA, 2008; 474p. [Google Scholar]
- Ruotsalainen, R.; Pukkala, T.; Kangas, A.; Vauhkonen, J.; Tuaominen, S. The effects of sample plot selection strategy and the number of sample plots on inoptimality losses in forest management planning based on airborne laser scanning data. Can. J. For. Res. 2019, 49, 1135–1146. [Google Scholar] [CrossRef]
- Hogland, J.; Anderson, N.; Chung, W. New Geospatial Approaches for Efficiently Mapping Forest Biomass Logistics at High Resolution over Large Areas. ISPRS Int. J. Geo-Inf. 2018, 7, 156. [Google Scholar] [CrossRef] [Green Version]
- America’s Longleaf. Longleaf Pine Maintenance Condition Class Definitions. Available online: http://www.americaslongleaf.org/media/mjroaokz/final-lpc-maintenance-condition-class-metrics-oct-2014-high-res.pdf (accessed on 2 April 2020).
- U.S. Forest Service; Forest Inventory and Analysis (FIA). Database, U.S. Department of Agriculture, Forest Service; Northern Research Station: Saint Paul, MN, USA, 2019. Available online: https://apps.fs.usda.gov/fia/datamart/datamart.html (accessed on 6 February 2019).












| Classification | Label | Definition/Query | Proportion |
|---|---|---|---|
| DFT | Pine (2) | (P_BAH > O_BAH) and not Nonforest | 0.480 |
| Other (1) | (O_BAH > P_BAH) and not Nonforest | 0.340 | |
| Nonforest (0) | P_BAH + O_BAH < 2 m2 ha−1 | 0.180 | |
| LPP | Present | LP_BAH > 0 m2 ha−1 and not Nonforest | 0.168 |
| Source | Code | Resolution | Bands | Spectral and Texture Metrics | Season | ID |
|---|---|---|---|---|---|---|
| Landsat 8 | L | 30 m | 2–7 | Mean (Cell) | LDS | 1–6 |
| LGS | 7–12 | |||||
| LWS | 13–18 | |||||
| Standard Deviation (3 by 3 Cells) | LDS | 19–24 | ||||
| LGS | 25–30 | |||||
| LWS | 31–36 | |||||
| Sentinel 2 | S | 10 m | 2, 3, 4, and 8 | Mean (3 by 3 Cells) | LDS | 1–4 |
| LGS | 5–8 | |||||
| LWS | 9–12 | |||||
| Standard Deviation (5 by 5 Cells) | LDS | 13–16 | ||||
| LGS | 17–20 | |||||
| LWS | 21–24 | |||||
| Quantum Spatial | A | 0.6 m | 1–4 | Mean (61 by 61 Cells) | LDS | 1–4 |
| Standard deviation (61 by 61 Cells) | LDS | 5–8 |
| Response | Normalization | Predictors * | Train | OOB | AIC |
|---|---|---|---|---|---|
| DFT | EANR | L3, L16, L17, S2, S5, S7, S21, A1 | 0.068 | 0.235 | 86.217 |
| RAW | L8, L9, L16, L18, S11, S5 | 0.103 | 0.263 | 110.023 | |
| LPP | EANR | L2, L17, S3, S4, A2, A5 | 0.056 | 0.129 | 94.683 |
| RAW | L2, L13, S9, A6 | 0.095 | 0.148 | 113.617 | |
| EANR | L2, L8, L10, L11, L16, L17, L18, L22, A2, A8 | 0.909 | 1.168 | 548.664 | |
| RAW | L2, L5, L6, L8, L10, L11, L17, L22, S9, S10, S11, S12, A4 | 0.900 | 1.267 | 547.441 | |
| EANR | L3, L5, L11, S7, S9, S21, S23, A2, A5 | 1.108 | 1.365 | 600.177 | |
| RAW | L3, L5, L11, L13, L16, S12, A5, A7 | 1.188 | 1.541 | 637.448 | |
| EANR | L3, L5, L8, L10, L11, L17, L28, S4, S11, A2, A5 | 5.755 | 9.197 | 1243.537 | |
| RAW | L1, L4, L5, l6, L13, L22, L31, S11, S20 | 6.709 | 10.866 | 1277.418 | |
| EANR | L9, L13, S2, S6, S7, S20, A1 | 11.787 | 14.977 | 1471.036 | |
| RAW | L3, L5, L11, L13, S12, S20, A5 | 12.492 | 14.682 | 1486.281 |
| EGAM * | GMI | p-Value |
|---|---|---|
| DFT (Nonforest) | 0.364 | <0.001 |
| DFT (Other) | 0.067 | 0.190 |
| DFT (Pine) | 0.093 | 0.153 |
| LPP (Present) | 0.223 | 0.002 |
| Pine BAH | 0.184 | 0.011 |
| Pine TPH | 0.068 | 0.188 |
| Other BAH | 0.087 | 0.131 |
| Other TPH | 0.140 | 0.037 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hogland, J.; Affleck, D.L.R.; Anderson, N.; Seielstad, C.; Dobrowski, S.; Graham, J.; Smith, R. Estimating Forest Characteristics for Longleaf Pine Restoration Using Normalized Remotely Sensed Imagery in Florida USA. Forests 2020, 11, 426. https://doi.org/10.3390/f11040426
Hogland J, Affleck DLR, Anderson N, Seielstad C, Dobrowski S, Graham J, Smith R. Estimating Forest Characteristics for Longleaf Pine Restoration Using Normalized Remotely Sensed Imagery in Florida USA. Forests. 2020; 11(4):426. https://doi.org/10.3390/f11040426
Chicago/Turabian StyleHogland, John, David L.R. Affleck, Nathaniel Anderson, Carl Seielstad, Solomon Dobrowski, Jon Graham, and Robert Smith. 2020. "Estimating Forest Characteristics for Longleaf Pine Restoration Using Normalized Remotely Sensed Imagery in Florida USA" Forests 11, no. 4: 426. https://doi.org/10.3390/f11040426