Discovery and Profiling of microRNAs at the Critical Period of Sex Differentiation in Xanthoceras sorbifolium Bunge
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Critical Period Determination
2.3. RNA Extraction, Libraries Construction, and High-Throughput Sequencing
2.4. Bioinformatics Analysis
2.5. Validation of Differentially Expressed miRNAs with RT-qPCR
3. Results
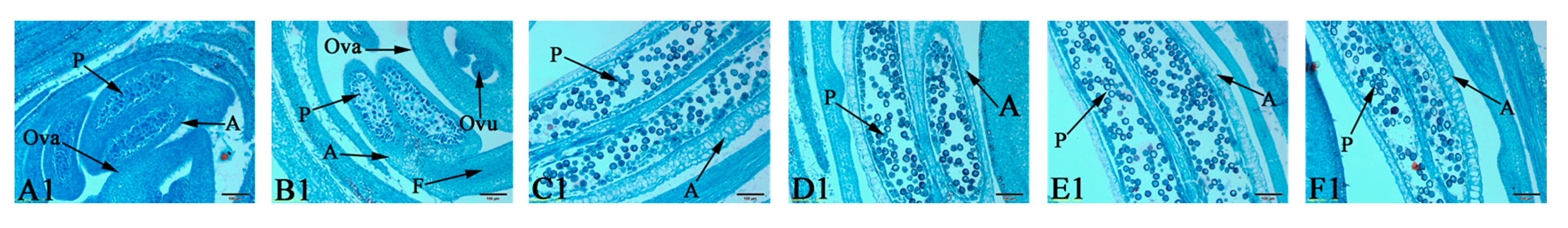
3.1. Determination of the Critical Period of Sex Differentiation
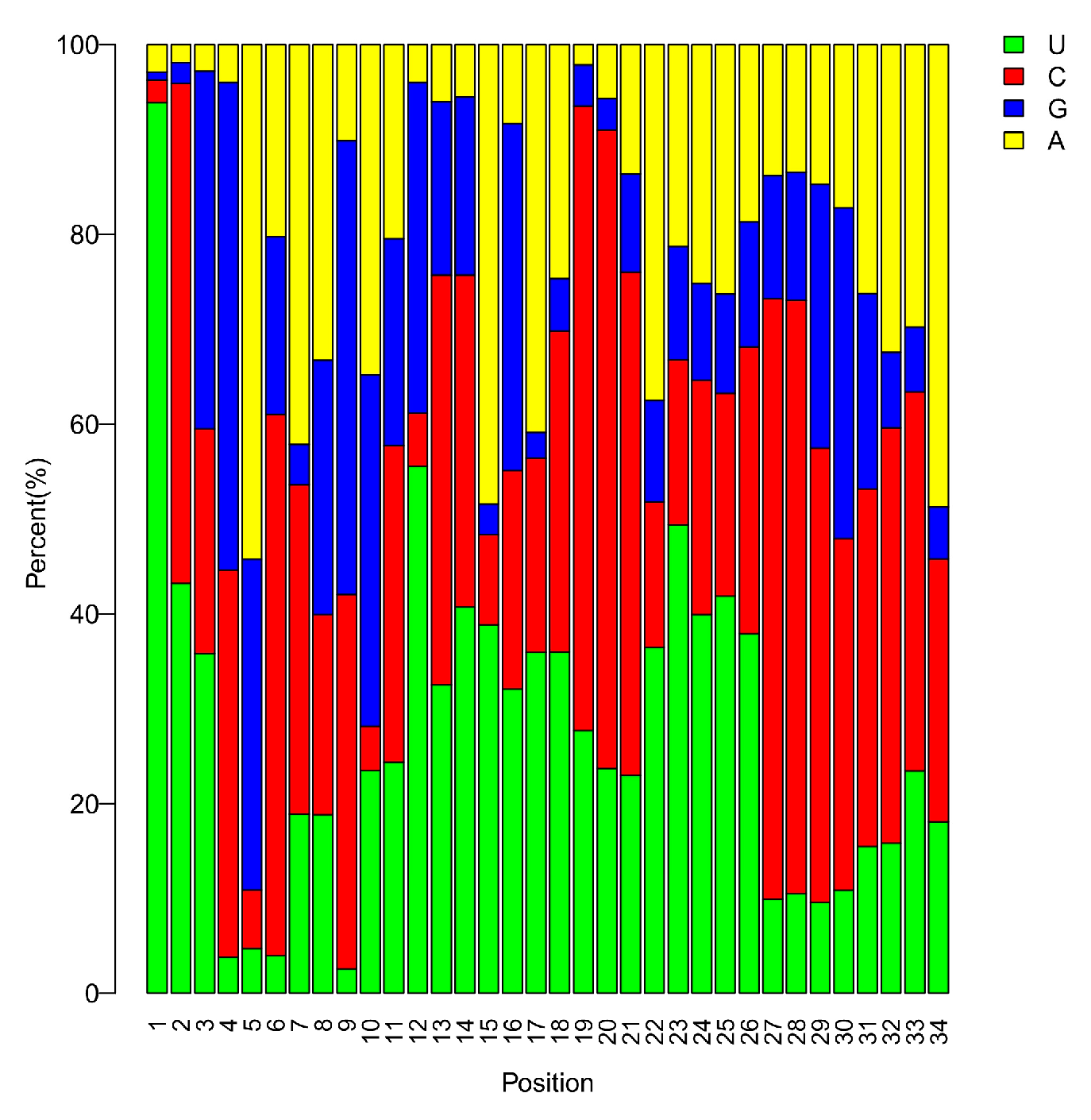
3.2. sRNA Profiles in the Critical Period of Sex Differentiation
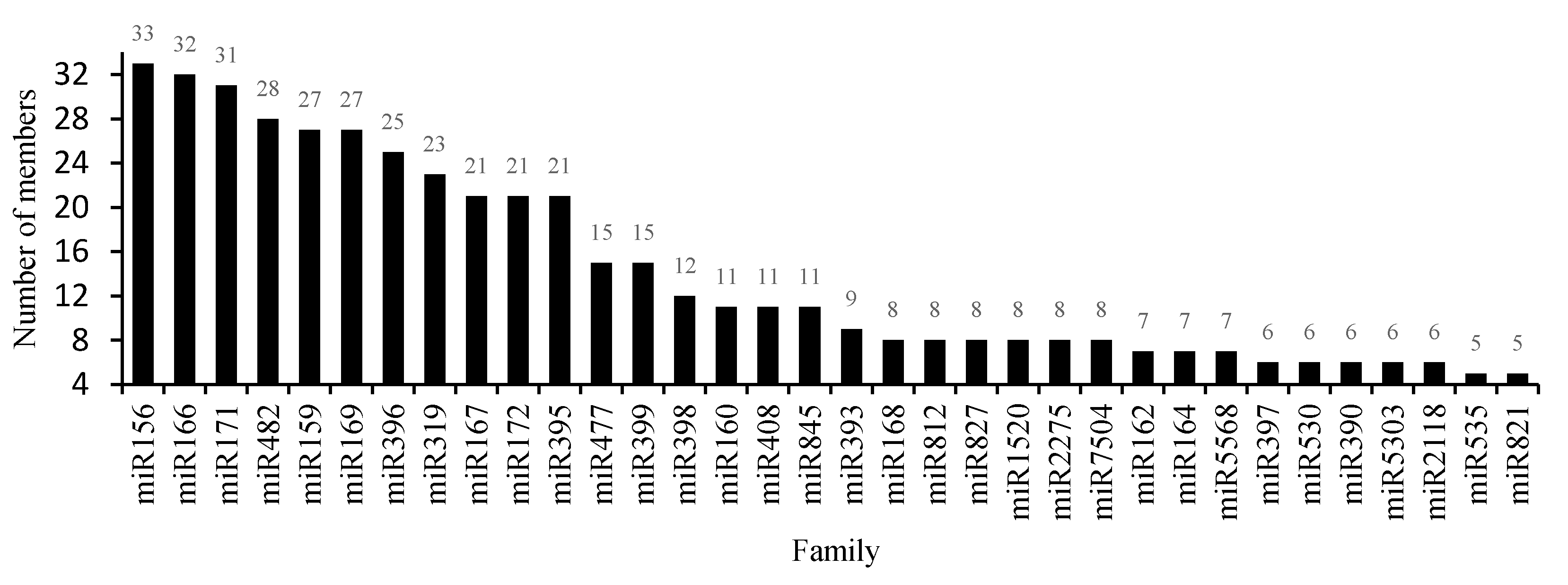
3.3. Conserved miRNAs in the Critical Period of Sex Differentiation
3.4. Novel miRNAs in the Critical Period of Sex Differentiation
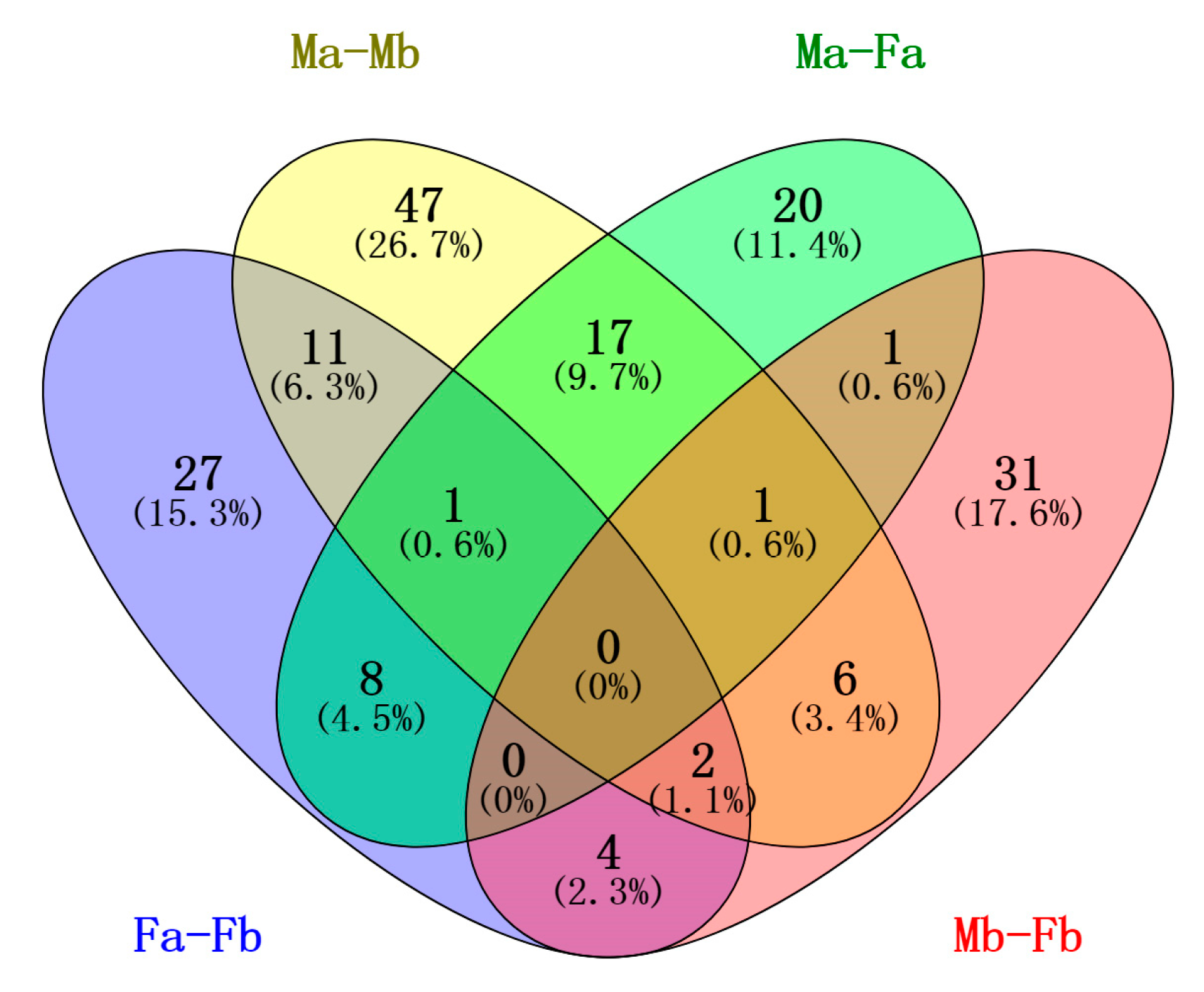
3.5. Differentially Expressed miRNAs among the Four Libraries
3.6. Target Prediction and Functional Annotation
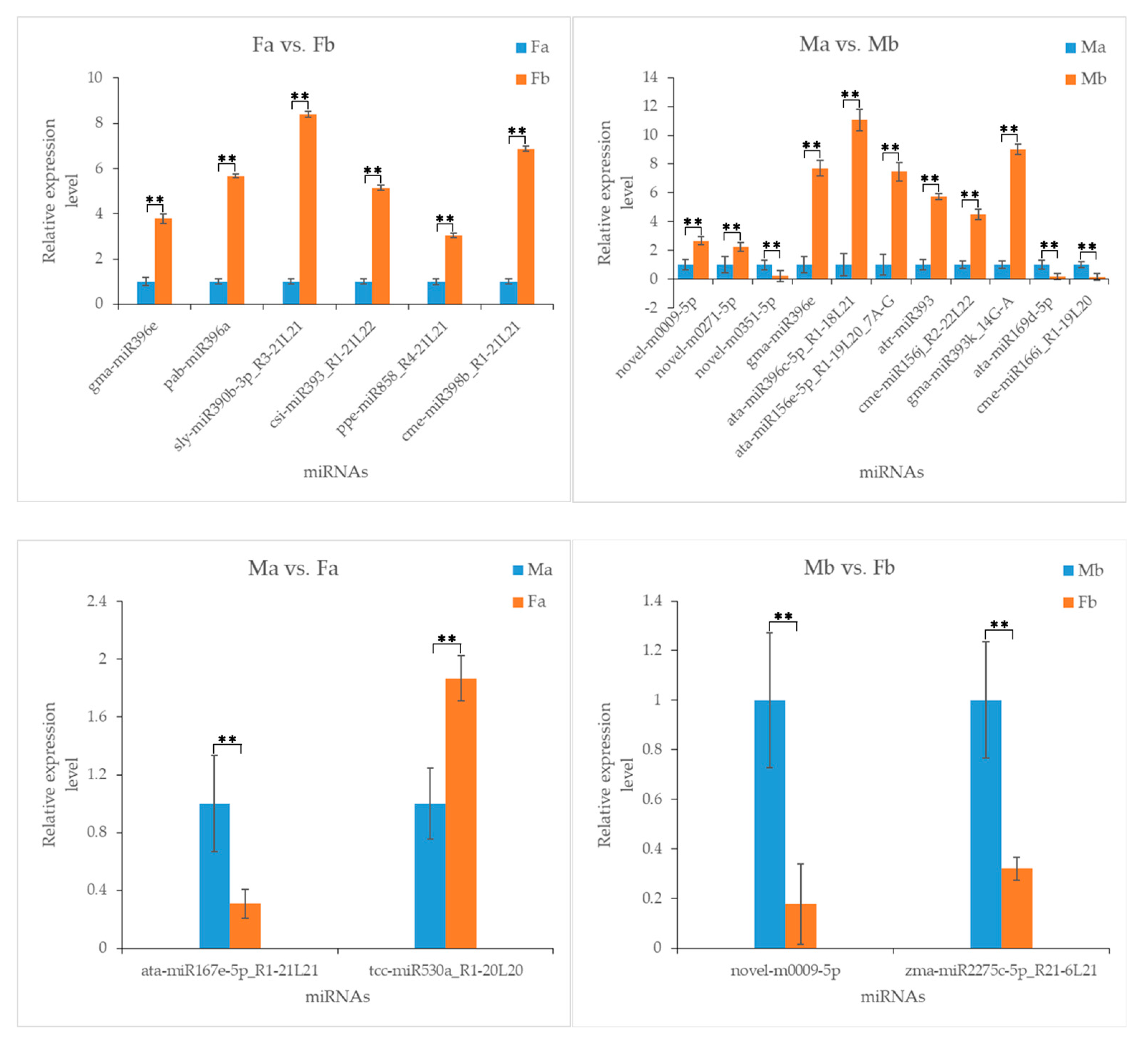
3.7. Validation of miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yu, H.Y.; Fan, S.Q.; Bi, Q.X.; Wang, S.X.; Hu, X.Y.; Chen, M.Y.; Wang, L.B. Seed morphology, oil content and fatty acid composition variability assessment in yellow horn (Xanthoceras sorbifolium Bunge) germplasm for optimum biodiesel production. Ind. Crops Prod. 2017, 97, 425–430. [Google Scholar] [CrossRef]
- Qi, J.H.; Yao, Z.Y. Review on reproductive biology, propagation and breeding of Xanthoceras sorbifolia. J. Northwest For. Univ. 2012, 27, 91–96. [Google Scholar]
- Li, J.; Zu, Y.G.; Fu, Y.J.; Yang, Y.C.; Li, S.M.; Li, Z.N.; Wink, M. Optimization of microwave-assisted extraction of triterpene saponins from defatted residue of yellow horn (Xanthoceras sorbifolia Bunge.) kernel and evaluation of its antioxidant activity. Innov. Food Sci. Emerg. Technol. 2010, 11, 637–643. [Google Scholar] [CrossRef]
- Li, Y.J.; Xu, J.K.; Xu, P.; Song, S.; Liu, P.; Chi, T.; Ji, X.; Jin, G.; Qiu, S.; Hou, Y.; et al. Xanthoceras sorbifolia extracts ameliorate dendritic spine deficiency and cognitive decline via upregulation of BDNF expression in a rat model of Alzheimer’s disease. Neurosci. Lett. 2016, 629, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.B.; Chu, L.Y. Resource evaluation of typical energy plants and possible functional zone planning in China. Biomass Bioenerg. 2008, 32, 283–288. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, K.Q.; Ao, Y.; Ma, L.Y.; Duan, J. Evaluation of biodiesel from Xanthoceras sorbifolia Bunge seed kernel oil from 13 areas in China. J. For. Res. 2019, 30, 869–877. [Google Scholar] [CrossRef]
- Ao, Y. Comparison of floral ontogeny between wild-type Xanthoceras sorbifolia bunge and its double-flowered mutant. Bangl. J. Bot. 2016, 45, 367–375. [Google Scholar]
- Wang, D.; Su, D.; Yu, B.; Chen, C.M.; Cheng, L.; Li, X.Z.; Xi, R.G.; Gao, H.Y.; Wang, X.B. Novel anti-tumour barringenol-like triterpenoids from the husks of Xanthoceras sorbifolia Bunge and their three dimensional quantitative structure activity relationships analysis. Fitoterapia 2017, 116, 51–60. [Google Scholar] [CrossRef]
- Kong, W.B.; Liang, J.Y.; Ma, Z.X.; Zhang, J. Research advance of Xanthoceras sorbifolia Bunge oil. China Oils Fats 2011, 36, 67–72. [Google Scholar]
- Harrington, K.J. Chemical and physical properties of vegetable oil esters and their effect on diesel fuel performance. Biomass 1986, 9, 1–17. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Zheng, Y.R. Comparative de novo transcriptome analysis of fertilized ovules in Xanthoceras sorbifolium uncovered a pool of genes expressed specifically or preferentially in the selfed ovule that are potentially involved in late-acting self-incompatibility. PLoS ONE 2015, 10, 0140507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, M.X.; Ao, Y. Research progress on the flowering and fruit set of Xanthoceras sorbifolia Bunge. Chin. Agric. Sci. Bull. 2008, 24, 381–384. [Google Scholar]
- Zhou, Q.Y.; Fu, D.Z. Preliminary studies on the seproductive siology of Xanthoceras sorbifolia. Sci. Silvae Sin. 2010, 46, 158–164. [Google Scholar]
- Zhang, N.; Ao, Y.; Su, S.C.; Liu, J.F.; Huang, Y.Y.; Liu, J.F.; Zhang, X.J. Analysis of morphological and anatomical features and meteorological factors during the sex differentiation in Bunge. Acta Bot. Boreali-Occident. Sin. 2018, 38, 86–97. [Google Scholar]
- Lü, X.Q.; Zhang, M.; Wang, D.; Wang, L. Comparative study on the bisexual flower and unisexual male flower of Bunge. Bull. Bot. Res. 2014, 34, 85–94. [Google Scholar]
- Ao, Y.; Duan, J.; Yu, H.Y.; Jiang, C.Y.; Ma, L.Y. Research progress on Xanthoceras sorbifolia Bunge. J. China Agric. Univ. 2012, 17, 197–203. [Google Scholar]
- Zhou, Q.Y.; Liu, G.S. The embryology of Xanthoceras and its phylogenetic implications. Plant Syst. Evol. 2012, 298, 457–468. [Google Scholar] [CrossRef]
- Hu, Q.; Gao, S.M.; Li, F.L. Pistil development in 2 types of flowers of Xanthoceras sorbifolia. For. Study China 2004, 6, 13–16. [Google Scholar] [CrossRef]
- Delong, A.; Calderonurrea, A.; Dellaporta, S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 1993, 74, 757–768. [Google Scholar] [CrossRef]
- Filipowicz, W.; Jaskiewicz, L.; Kolb, F.A.; Pillai, R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struc. Biol. 2005, 15, 331–341. [Google Scholar] [CrossRef]
- Chen, X.M. Small RNAs—secrets and surprises of the genome. Plant J. 2010, 61, 941–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campo, S.; Peris-Peris, C.; Siré, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; Segundo, B.S. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. Micrornas in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.X.; Pan, T.F.; Pan, D.M. Research advances on regulation of microRNAs on floral bud differentiation. J. Anhui Agri. Sci. 2014, 42, 9682–9683. [Google Scholar]
- Zhang, Q.; Li, J.H.; Sang, Y.L.; Xing, S.Y.; Wu, Q.K.; Liu, X.J. Identification and characterization of microRNAs in Ginkgo biloba var. epiphylla Mak. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, O.P.; Sharma, P.; Gupta, R.K.; Sharma, I. MicroRNA mediated regulation of metal toxicity in plants: Present status and future perspectives. Plant Mol. Biol. 2014, 84, 1–18. [Google Scholar] [CrossRef]
- Allen, R.S.; Li, J.Y.; Stahle, M.I.; Dubroué, A.; Gubler, F.; Millar, A.A. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 2007, 104, 16371–16376. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.L.; Zheng, Y.; Qin, L.; Wang, Y.; Chen, L.F.; He, Y.J.; Fei, Z.J.; Lu, G. Identification of miRNAs and their targets through high-throughput sequencing and degradome analysis in male and female Asparagus officinalis. BMC Plant Biol. 2016, 16, 80. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Park, M.Y.; Conway, S.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [Green Version]
- Li, X.B.; Jin, F.; Jin, L.; Jackson, A.; Ma, X.; Shu, X.L.; Wu, D.X.; Jin, G.Q. Characterization and comparative profiling of the small RNA transcriptomes in two phases of flowering in Cymbidium ensifolium. BMC Genom. 2015, 16, 622. [Google Scholar] [CrossRef] [Green Version]
- Banks, J.A. MicroRNA, sex determination and floral meristem determinacy in maize. Genome Biol. 2008, 9, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Le Trionnaire, G.; Twell, D. Small RNAs in angiosperm gametophytes: From epigenetics to gamete development. Genes Dev. 2010, 24, 1081–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ao, Y.; Wang, Y.W.; Chen, L.; Wang, T.; Yu, H.Y.; Zhang, Z.X. Identification and comparative profiling of microRNAs in wild-type Xanthoceras sorbifolia and its double flower mutant. Genes Genom. 2012, 34, 561–568. [Google Scholar] [CrossRef]
- Bi, Q.X.; Guo, B.; Zhang, D.X.; Guan, W.B. Identification and characterization of conserved and novel microRNAs in Xanthoceras sorbifolium via deep sequencing. Genes Genom. 2015, 37, 281–286. [Google Scholar] [CrossRef]
- Ao, Y. Characterization and comparison of flower bud microRNAs from yellow-horn species. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rfam. Available online: http://rfam.janelia.org/ (accessed on 21 October 2019).
- miRBase. Available online: ftp://mirbase.org/pub/mirbase/CURRENT/ (accessed on 21 October 2019).
- Reference genome. Available online: ftp://ftp.ensembl.org/pub/release-85/fasta/homo_sapiens/dna/Homo_sapiens.GRCh38.dna.toplevel.fa.gz (accessed on 21 October 2019).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.; Green, P.J.; et al. Criteria for Annotation of Plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef]
- Enright, A.J.; Bino, J.; Ulrike, G. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GO. Available online: http://geneontology.org/ (accessed on 21 October 2019).
- KEGG. Available online: http://www.genome.jp/kegg (accessed on 21 October 2019).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Du, X.H. Study on molecular mechanism of male sterility in Xanthoceras Sorbifolium Bunge. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2003. [Google Scholar]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, R.D.; Aksay, G.; Dolgosheina, E.; Ebhardt, H.A.; Magrini, V.; Mardis, E.R.; Sahinalp, S.C.; Unrau, P.J. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008, 18, 571–584. [Google Scholar] [CrossRef] [Green Version]
- Mao, W.H.; Li, Z.Y.; Xia, X.J.; Li, Y.D.; Yu, J.Q. A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS ONE 2012, 7, 33040. [Google Scholar] [CrossRef] [Green Version]
- Song, C.N.; Wang, C.; Zhang, C.Q.; Korir, N.K.; Yu, H.P.; Ma, Z.Q.; Fang, J.G. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom. 2010, 11, 431. [Google Scholar] [CrossRef] [Green Version]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and Functional Diversification of MIRNA Genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.Z.; Dong, C.; Wang, G.; Zheng, X.W.; Li, W.C. Advances in floral bud differentiation and floral sex determination in litchi. Guangdong Agric. Sci. 2017, 44, 34–40. [Google Scholar]
- Wang, P.; Zheng, W.; Chen, W. Fluorescence microscopic observation on flower sex differentiation in Litchi (Litchi chinensis Sonn.). Chin. J. Trop. Crops 2010, 31, 740–744. [Google Scholar]
- Wang, H.G. Study on the cytology mechanism of flower sexual differentiation in Longan (Dimocarpus longan Lour.). Master’s Thesis, Fujian Agriculture Forestry University, Fuzhou, China, 2008. [Google Scholar]
- Wang, C.C.; Ke, G.W. An observation on the inflorescence development and the sequence of flower differentiation of Longan. Fujian J. Agric. Sci. 1988, 8, 434–436. [Google Scholar]
- Mo, C.M.; Tu, D.P.; Huang, J. Morphological and endogenous hormones characteristics of flower bud of siraitia grosvenorii during its differentiation. Acta Bot. Boreali-Occident. Sin. 2015, 35, 98–106. [Google Scholar]
- Yuan, G.F.; Zhao, Q.P.; Sun, H.Y.; Wang, Q.M. Micromorphlogical analysis of sex organ development and SDS-PAGE profile of male flowers at their later developmental stages in cucumber (Cucumis sativus L.). J. Zhejiang Univ. Agric. Life Sci. 2005, 31, 145–150. [Google Scholar]
- Hong, Y.G.; Jackson, S. Floral induction and flower formation-the role and potential applications of miRNAs. Plant Biotechnol. J. 2015, 13, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.M.; Yang, F.X.; Yu, D.Q. Overexpression of miR396 miRNAs caused Flower stigma curved in Arabidopsis thaliana. Acta Bot. Yunnanica 2009, 31, 353–356. [Google Scholar] [CrossRef]
- Ma, C.; Burd, S.; Lers, A. miR408, is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Inês, T.; Cláudio, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M.D. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar]
- Xing, S.P.; Salinas, M.; Garcia-Molina, A.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 2013, 75, 566–577. [Google Scholar] [CrossRef]
- Xing, S.P.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.H.; Bao, M.L.; Sun, Y.Z.; Yang, Y.J.; Xu, X.H.; Wang, J.H.; Han, N.; Bian, H.W.; Zhu, M.Y. Regulation of auxin response by miR393-targetedtransport inhibitor response protein1is involved in normal development in Arabidopsis. Plant Mol. Biol. 2011, 77, 619–629. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongué, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, 107678. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, Y.; Chen, H.Y. The effect of low night temperature on miRNA expression in mutant and wild type tomato leaf. Acta Hortic. Sin. 2016, 43, 2369–2379. [Google Scholar]
- Alonso-Peral, M.M.; Li, J.; Li, Y.; Allen, R.S.; Schnippenkoetter, W.; Ohms, S.; White, R.G.; Millar, A.A. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154, 757–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gocal, G.F.W.; Sheldon, C.C.; Gubler, F.; Moritz, T.; Bagnall, D.J.; MacMillan, C.P.; Li, S.F.; Parish, R.W.; Dennis, E.S.; Weigel, D.; et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 2001, 127, 1682–1693. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schommer, C.; Bresso, E.G.; Spinelli, S.V.; Palatnik, J.F. Role of MicroRNA miR319 in plant development. Signal. Commun. Plants 2012, 15, 29–47. [Google Scholar]







| miRNA | Putative Target Gene | Target Gene Function |
|---|---|---|
| gma-miR393k_14G-A | TR6435|c4_g4 | Pollen maturation; Stamen development |
| mes-miR393d | ||
| atr-miR393 | ||
| gma-miR396e | TR3538|c0_g1 | Embryo sac development; Ovule development |
| ata-miR396c-5p_R1-18L21 | ||
| ata-miR396e-3p_R1-15L21 | ||
| cme-miR159b_R2-21L21 | TR1924|c0_g1 | Flower development |
| gma-miR319l | ||
| gma-miR319q | ||
| cme-miR156j_R2-22L22 | TR12296|c0_g1 | Anthers development |
| ata-miR156e-5p_R1-19L20_7A-G | ||
| ata-miR156e-5p_R1-19L20_7A-G | TR10002|c0_g1 | Pollen tube development |
| TR3055|c0_g1 | Flower development; Phyllotaxis development | |
| ata-miR408-3p_R1-17L20 | TR16019|c0_g1 | Embryo development; Flower development; Fruit development; Leaf development |
| TR10727|c0_g1 | Organ growth | |
| bra-miR162-3p_R1-17L21 | TR9505|c0_g1 | Embryo sac development |
| mtr-miR2673b_R17-2L22 | TR1482|c0_g1 | Pollen maturation |
| bdi-miR5169b_R7-21L21 | TR4178|c0_g1 | Flower development |
| novel-m0351-5p | TR8265|c0_g1 | Anthers development |
| novel-m0318-5p | TR10202|c0_g1 | Flower development regulation; Flower organ formation |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zheng, Y.; Su, S.; Ao, Y. Discovery and Profiling of microRNAs at the Critical Period of Sex Differentiation in Xanthoceras sorbifolium Bunge. Forests 2019, 10, 1141. https://doi.org/10.3390/f10121141
Wang X, Zheng Y, Su S, Ao Y. Discovery and Profiling of microRNAs at the Critical Period of Sex Differentiation in Xanthoceras sorbifolium Bunge. Forests. 2019; 10(12):1141. https://doi.org/10.3390/f10121141
Chicago/Turabian StyleWang, Xu, Yaqi Zheng, Shuchai Su, and Yan Ao. 2019. "Discovery and Profiling of microRNAs at the Critical Period of Sex Differentiation in Xanthoceras sorbifolium Bunge" Forests 10, no. 12: 1141. https://doi.org/10.3390/f10121141





