Adhesion and Growth of Neuralized Mouse Embryonic Stem Cells on Parylene-C/SiO2 Substrates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Maintenance and Culture of Mouse Embryonic Stem Cells
2.3. Embryonic Stem Cell Neuralisation
2.4. Primary Cell Culture
2.5. Immunocytochemistry and Cell Labeling
2.6. Pattern Fabrication, Sterilisation and Activation
2.7. Statistical Analysis
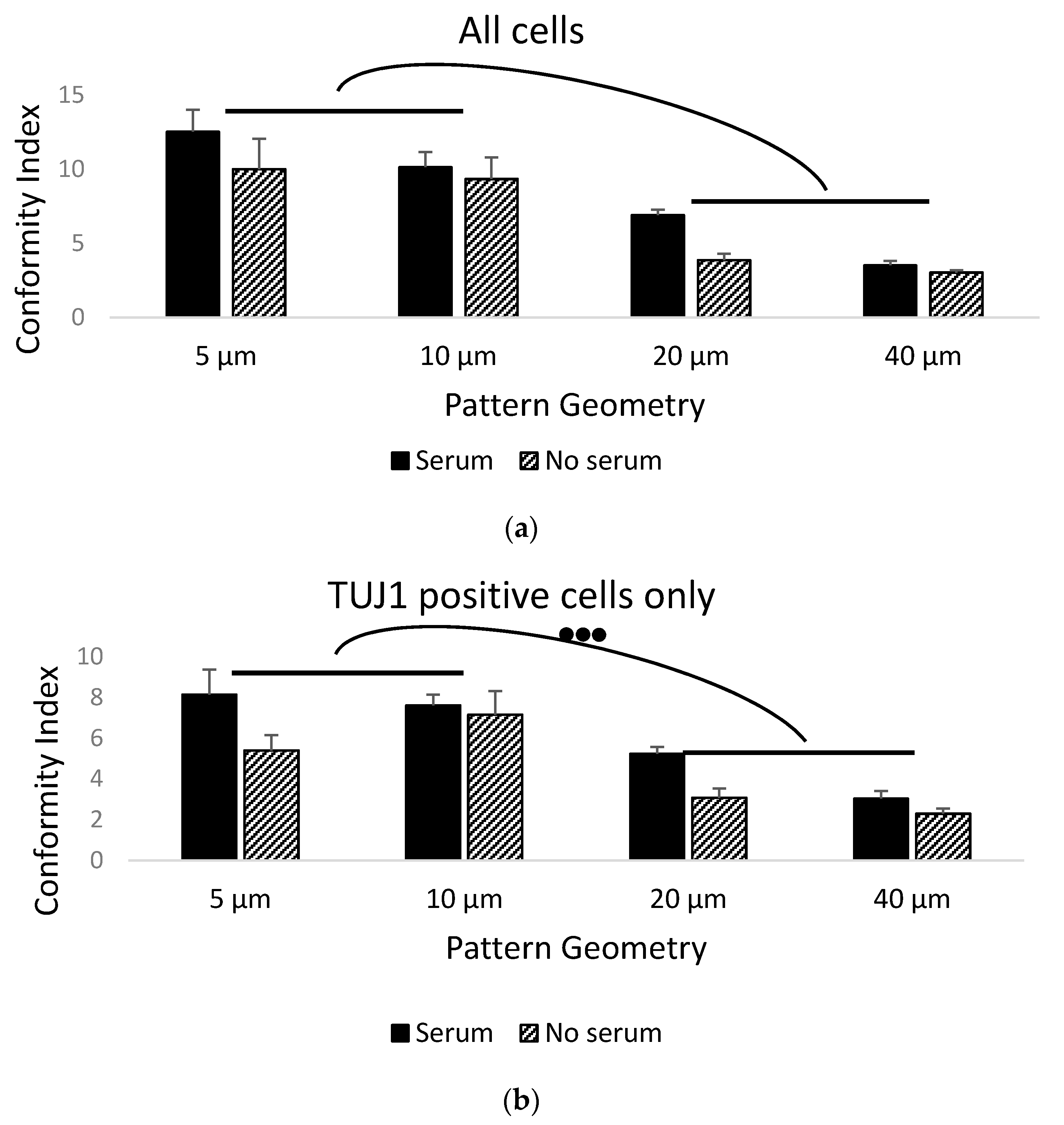
3. Results
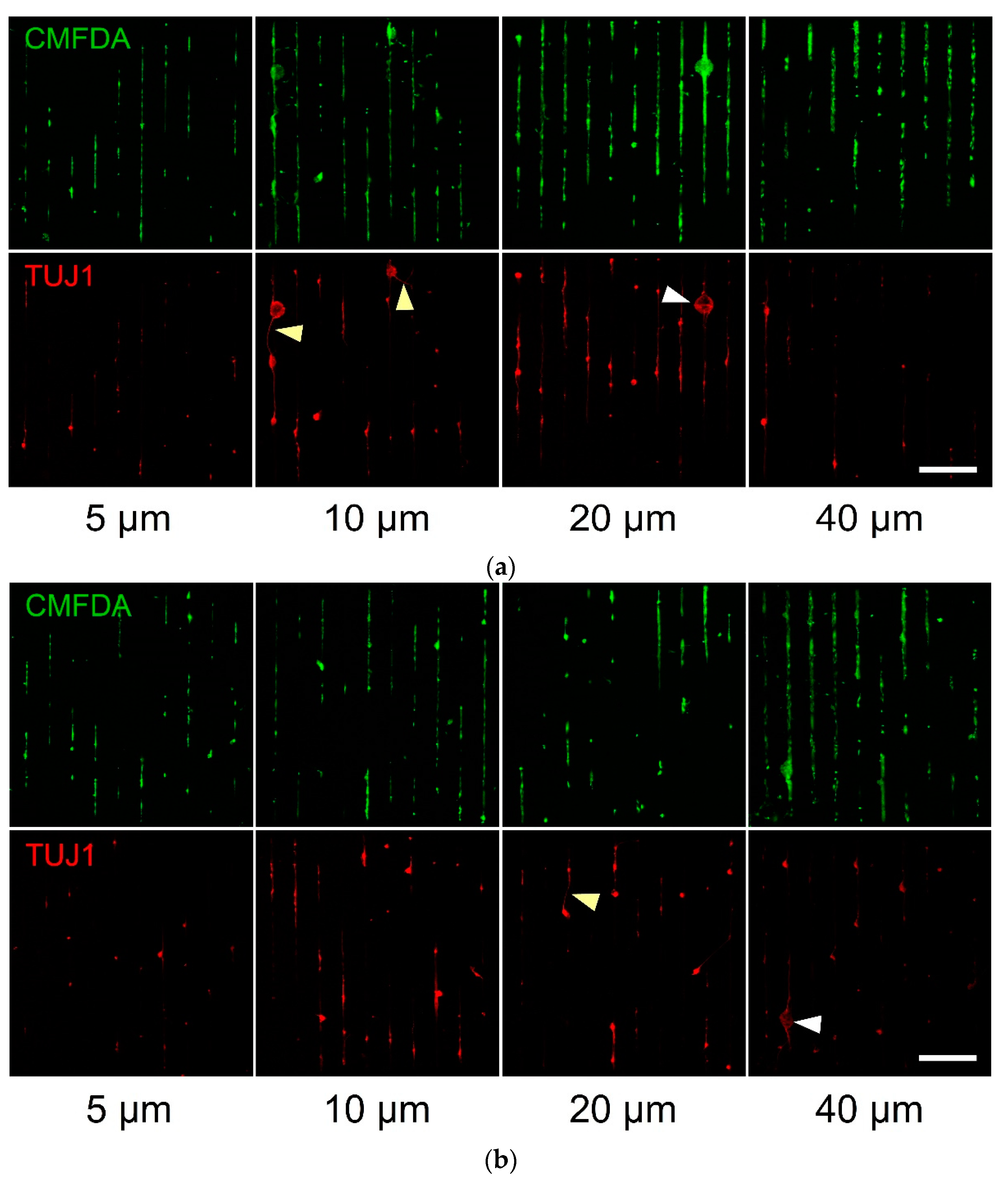
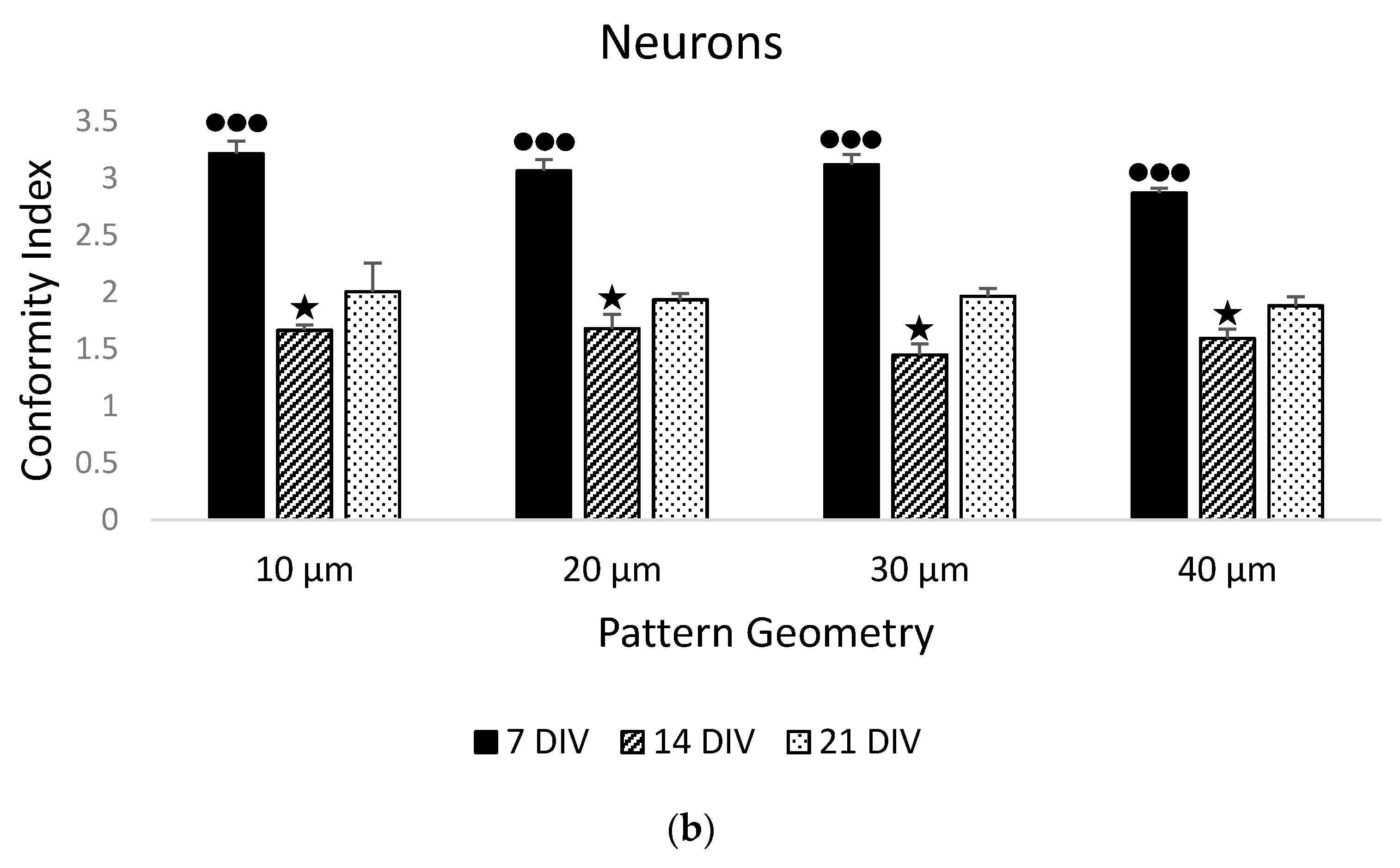
3.1. Stem Cell Guidance on Parylene-C Patterns
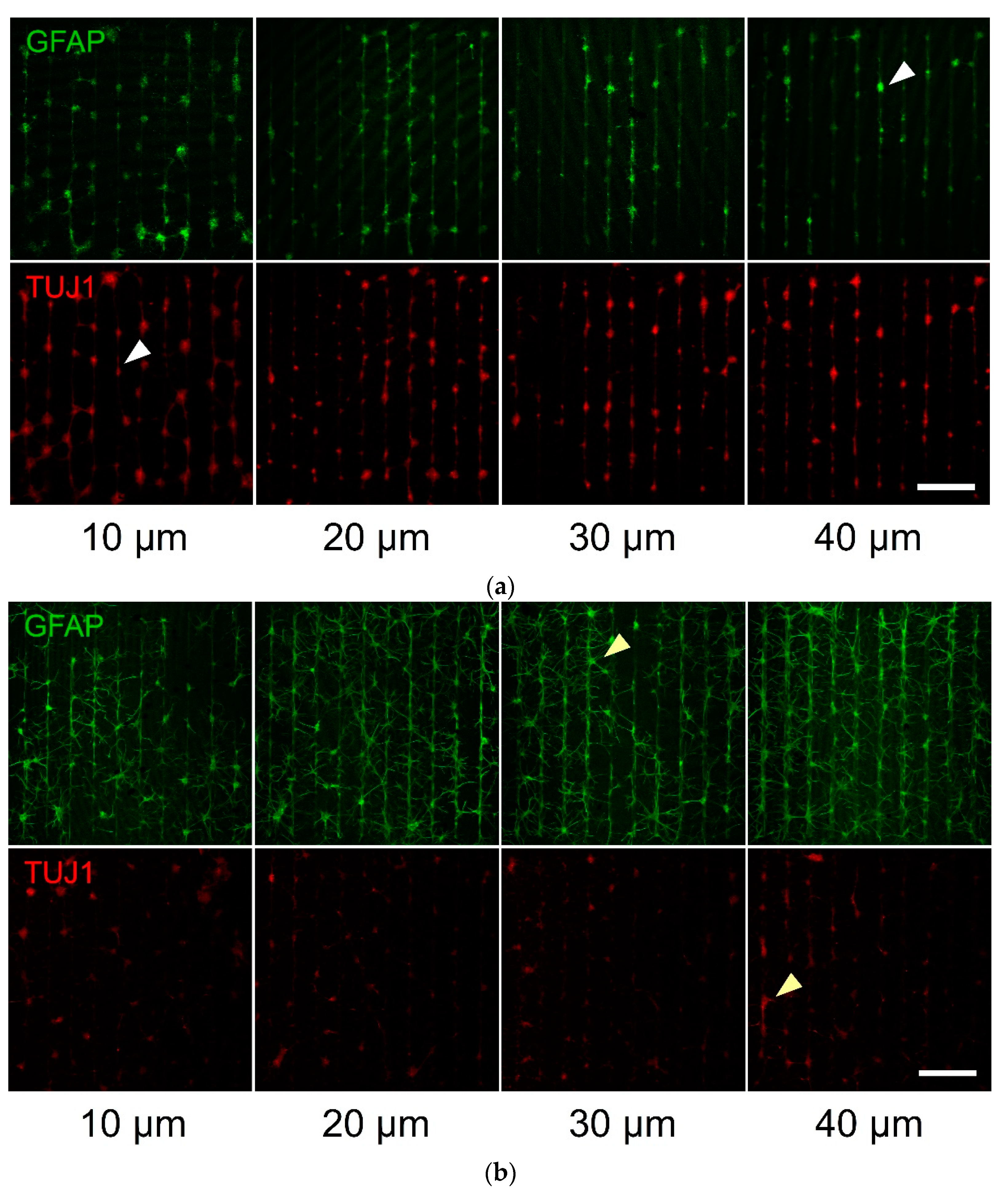
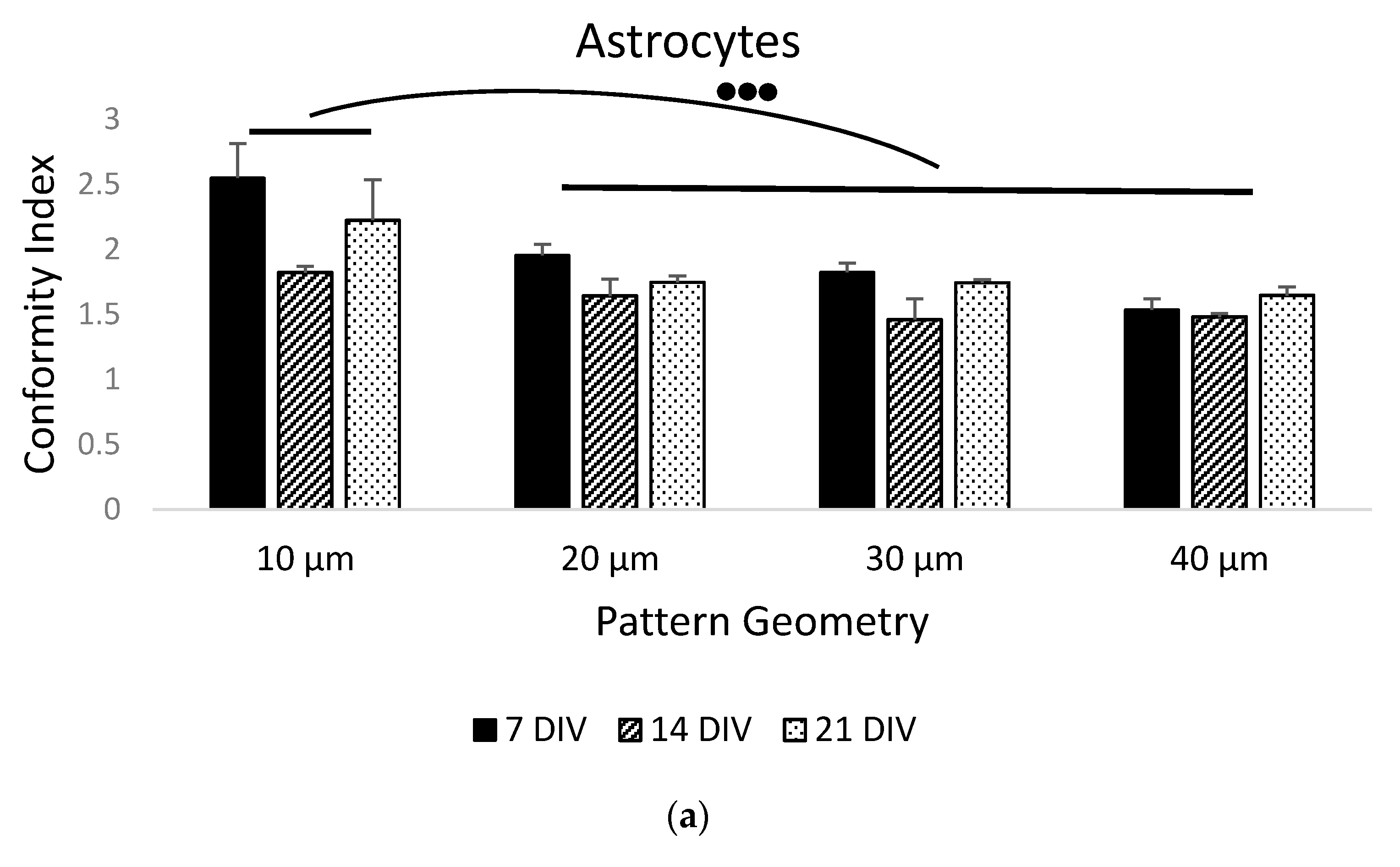
3.2. Primary Cell Patterning with Parylene-C
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bang, S.; Na, S.; Jang, J.M.; Kim, J.; Jeon, N.L. Engineering-Aligned 3D Neural Circuit in Microfluidic Device. Adv. Healthc. Mater. 2016, 5, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Samhaber, R.; Schottdorf, M.; El Hady, A.; Bröking, K.; Daus, A.; Thielemann, C.; Stühmer, W.; Wolf, F. Growing neuronal islands on multi-electrode arrays using an accurate positioning-μCP device. J. Neurosci. Methods 2016, 257, 194–203. [Google Scholar] [CrossRef]
- Li, W.; Xu, Z.; Huang, J.; Lin, X.; Luo, R.; Chen, C.H.; Shi, P. NeuroArray: A universal interface for patterning and interrogating neural circuitry with single cell resolution. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahnavi, S.; Arthi, N.; Pallavi, S.; Selvaraju, C.; Bhuvaneshwar, G.S.; Kumary, T.V.; Verma, R.S. Nanosecond laser ablation enhances cellular infiltration in a hybrid tissue scaffold. Mater. Sci. Eng. C 2017, 77, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Palyvoda, O.; Chen, C.C.; Auner, G.W. Culturing neuron cells on electrode with self-assembly monolayer. Biosens. Bioelectron. 2007, 22, 2346–2350. [Google Scholar] [CrossRef]
- Fannon, O.M.; Bithell, A.; Whalley, B.J.; Delivopoulos, E. A Fiber Alginate Co-culture Platform for the Differentiation of mESC and Modeling of the Neural Tube. Front. Neurosci. 2021, 14, 1386. [Google Scholar] [CrossRef]
- Faustino Martins, J.M.; Fischer, C.; Urzi, A.; Vidal, R.; Kunz, S.; Ruffault, P.L.; Kabuss, L.; Hube, I.; Gazzerro, E.; Birchmeier, C.; et al. Self-Organizing 3D Human Trunk Neuromuscular Organoids. Cell Stem Cell 2020, 26, 172–186.e6. [Google Scholar] [CrossRef] [PubMed]
- Specht, C.G.; Williams, O.A.; Jackman, R.B.; Schoepfer, R. Ordered growth of neurons on diamond. Biomaterials 2004, 25, 4073–4078. [Google Scholar] [CrossRef]
- Gu, Q.; Tomaskovic-Crook, E.; Lozano, R.; Chen, Y.; Kapsa, R.M.; Zhou, Q.; Wallace, G.G.; Crook, J.M. Functional 3D Neural Mini-Tissues from Printed Gel-Based Bioink and Human Neural Stem Cells. Adv. Healthc. Mater. 2016, 5, 1429–1438. [Google Scholar] [CrossRef] [Green Version]
- Castagnola, V.; Descamps, E.; Lecestre, A.; Dahan, L.; Remaud, J.; Nowak, L.G.; Bergaud, C. Parylene-based flexible neural probes with PEDOT coated surface for brain stimulation and recording. Biosens. Bioelectron. 2015, 67, 450–457. [Google Scholar] [CrossRef]
- Kang, Y.N.; Chou, N.; Jang, J.W.; Byun, D.; Kang, H.; Moon, D.J.; Kim, J.; Kim, S. An Intrafascicular Neural Interface with Enhanced Interconnection for Recording of Peripheral Nerve Signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1312–1319. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wolf, N.; Hondrich, T.J.J.; Shokoohimehr, P.; Milos, F.; Glass, M.; Mayer, D.; Maybeck, V.; Prömpers, M.; Offenhäusser, A.; et al. Engineering Biocompatible Interfaces via Combinations of Oxide Films and Organic Self-Assembled Monolayers. ACS Appl. Mater. Interfaces 2020, 12, 17121–17129. [Google Scholar] [CrossRef]
- Ramadi, K.B.; Dagdeviren, C.; Bhagchandani, P.; Nunez-Lopez, C.; Kim, M.J.; Langer, R.; Graybiel, A.M.; Cima, M.J. Simultaneous recording and marking of brain microstructures. J. Neural Eng. 2020, 17, 44001. [Google Scholar] [CrossRef]
- Delivopoulos, E.; Ouberai, M.M.; Coffey, P.D.; Swann, M.J.; Shakesheff, K.M.; Welland, M.E. Serum protein layers on parylene-C and silicon oxide: Effect on cell adhesion. Colloids Surf. B Biointerfaces 2015, 126, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Delivopoulos, E.; Murray, A.F.; MacLeod, N.K.; Curtis, J.C. Guided growth of neurons and glia using microfabricated patterns of parylene-C on a SiO2 background. Biomaterials 2009, 30, 2048–2058. [Google Scholar] [CrossRef]
- Raos, B.J.; Unsworth, C.P.; Costa, J.L.; Rohde, C.A.; Doyle, C.S.; Bunting, A.S.; Delivopoulos, E.; Murray, A.F.; Dickinson, M.E.; Simpson, M.C.; et al. Infra-red laser ablative micromachining of parylene-C on SiO2 substrates for rapid prototyping, high yield, human neuronal cell patterning. Biofabrication 2013, 5, 025006. [Google Scholar] [CrossRef]
- Unsworth, C.P.; Delivopoulos, E.; Gillespie, T.; Murray, A.F. Isolating single primary rat hippocampal neurons & astrocytes on ultra-thin patterned parylene-C/silicon dioxide substrates. Biomaterials 2011, 32, 2566–2574. [Google Scholar]
- Delivopoulos, E.; Murray, A.F.; Curtis, J.C. Effects of parylene-C photooxidation on serum-assisted glial and neuronal patterning. J. Biomed. Mater. Res. Part A 2010, 94, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Delivopoulos, E.; Murray, A.F. Controlled Adhesion and Growth of Long Term Glial and Neuronal Cultures on Parylene-C. PLoS ONE 2011, 6, e25411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unsworth, C.P.; Graham, E.S.; Delivopoulos, E.; Dragunow, M.; Murray, A.F. First human hNT neurons patterned on parylene-C/silicon dioxide substrates: Combining an accessible cell line and robust patterning technology for the study of the pathological adult human brain. J. Neurosci. Methods 2010, 194, 154–157. [Google Scholar] [CrossRef]
- Unsworth, C.P.; Holloway, H.; Delivopoulos, E.; Murray, A.F.; Simpson, M.C.; Dickinson, M.E.; Graham, E.S. Patterning and detailed study of human hNT astrocytes on parylene-C/silicon dioxide substrates to the single cell level. Biomaterials 2011, 32, 6541–6550. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.D.; Raos, B.J.; Bunting, A.S.; Murray, A.F.; Graham, E.S.; Unsworth, C.P. Human astrocytic grid networks patterned in parylene-C inlayed SiO2 trenches. Biomaterials 2016, 105, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Raos, B.J.; Simpson, M.C.; Doyle, C.S.; Murray, A.F.; Graham, E.S.; Unsworth, C.P. Patterning of functional human astrocytes onto parylene-C/SiO2 substrates for the study of Ca2+ dynamics in astrocytic networks. J. Neural Eng. 2018, 15, 036015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, D.; Rajalingam, B.; Selvarasah, S.; Dokmeci, M.R.; Khademhosseini, A. Generation of static and dynamic patterned co-cultures using microfabricated parylene-C stencils. Lab Chip 2007, 7, 1272–1279. [Google Scholar] [CrossRef]
- Jinno, S.; Moeller, H.-C.; Chen, C.-L.; Rajalingam, B.; Chung, B.G.; Dokmeci, M.R.; Khademhosseini, A. Microfabricated multilayer parylene-C stencils for the generation of patterned dynamic co-cultures. J. Biomed. Mater. Res. Part A 2008, 86A, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Lu, H.; Kawazoe, N.; Chen, G. Adipogenic differentiation of individual mesenchymal stem cell on different geometric micropatterns. Langmuir 2011, 27, 6155–6162. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Z.; Xie, C.; Wang, X.; Luo, F.; Hong, M.; Zhou, R.; Ma, C.; Lin, N.; Zhang, J.; et al. Scaffold with Micro/Macro-Architecture for Myocardial Alignment Engineering into Complex 3D Cell Patterns. Adv. Healthc. Mater. 2019, 8, 1901015. [Google Scholar] [CrossRef]
- Wang, X.; Nakamoto, T.; Dulińska-Molak, I.; Kawazoe, N.; Chen, G. Regulating the stemness of mesenchymal stem cells by tuning micropattern features. J. Mater. Chem. B 2016, 4, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, X.; Huang, T.C.; Hu, X.; Kawazoe, N.; Tsai, W.B.; Yang, Y.; Chen, G. Regulation of mesenchymal stem cell functions by micro-nano hybrid patterned surfaces. J. Mater. Chem. B 2018, 6, 5424–5434. [Google Scholar] [CrossRef]
- Han, S.; Kim, J.; Li, R.; Ma, A.; Kwan, V.; Luong, K.; Sohn, L.L. Hydrophobic Patterning-Based 3D Microfluidic Cell Culture Assay. Adv. Healthc. Mater. 2018, 7, e1800122. [Google Scholar] [CrossRef]
- Rosenthal, A.; Macdonald, A.; Voldman, J. Cell patterning chip for controlling the stem cell microenvironment. Biomaterials 2007, 28, 3208–3216. [Google Scholar] [CrossRef] [Green Version]
- Peljto, M.; Dasen, J.S.; Mazzoni, E.O.; Jessell, T.M.; Wichterle, H. Functional Diversity of ESC-Derived Motor Neuron Subtypes Revealed through Intraspinal Transplantation. Cell Stem Cell 2010, 7, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Brewer, G.J. Isolation and culture of adult rat hippocampal neurons. J. Neurosci. Methods 1997, 71, 143–155. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, Y.; Kokovay, E.; Lin, G.; Chuang, S.M.; Goderie, S.K.; Roysam, B.; Temple, S. Adult SVZ Stem Cells Lie in a Vascular Niche: A Quantitative Analysis of Niche Cell-Cell Interactions. Cell Stem Cell 2008, 3, 289–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gladson, C.L.; Stewart, J.E.; Olman, M.A.; Chang, P.L.; Schnapp, L.M.; Grammer, J.R.; Benveniste, E.N. Attachment of primary neonatal rat astrocytes to vitronectin is mediated by integrins αvβ5 and α8β1: Modulation by the type 1 plasminogen activator inhibitor. Neurosci. Lett. 2000, 283, 157–161. [Google Scholar] [CrossRef]
- Corey, J.M.; Wheeler, B.C.; Brewer, G.J. Micrometer resolution silane-based patterning of hippocampal neurons: Critical variables in photoresist and laser ablation processes for substrate fabrication. Biomed. Eng. IEEE Trans. 1996, 43, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Lauer, L.; Klein, C.; Offenhäusser, A. Spot compliant neuronal networks by structure optimized micro-contact printing. Biomaterials 2001, 22, 1925–1932. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murray, A.F.; Delivopoulos, E. Adhesion and Growth of Neuralized Mouse Embryonic Stem Cells on Parylene-C/SiO2 Substrates. Materials 2021, 14, 3174. https://doi.org/10.3390/ma14123174
Murray AF, Delivopoulos E. Adhesion and Growth of Neuralized Mouse Embryonic Stem Cells on Parylene-C/SiO2 Substrates. Materials. 2021; 14(12):3174. https://doi.org/10.3390/ma14123174
Chicago/Turabian StyleMurray, Alan F., and Evangelos Delivopoulos. 2021. "Adhesion and Growth of Neuralized Mouse Embryonic Stem Cells on Parylene-C/SiO2 Substrates" Materials 14, no. 12: 3174. https://doi.org/10.3390/ma14123174




