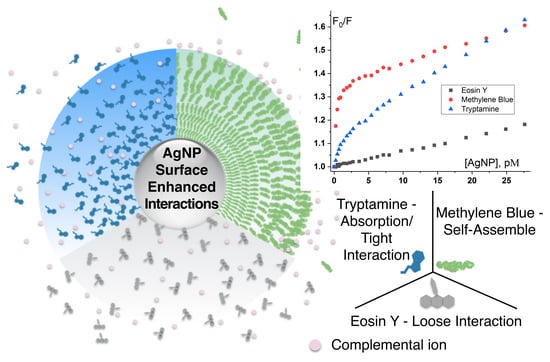
Silver Nanoparticle Surface Enabled Self-Assembly of Organic Dye Molecules
Abstract
:1. Introduction
2. Methods
3. Results and Discussion
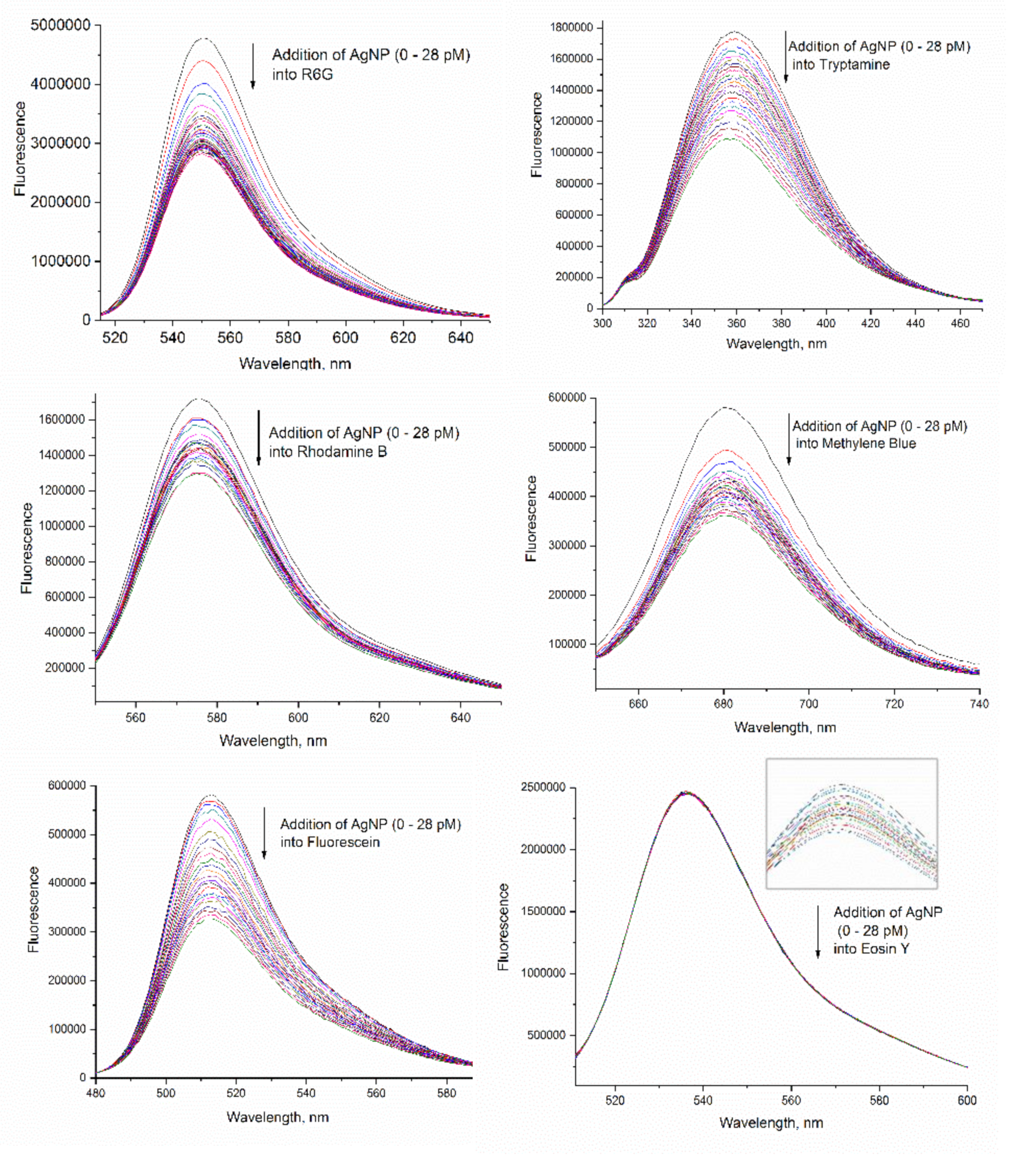
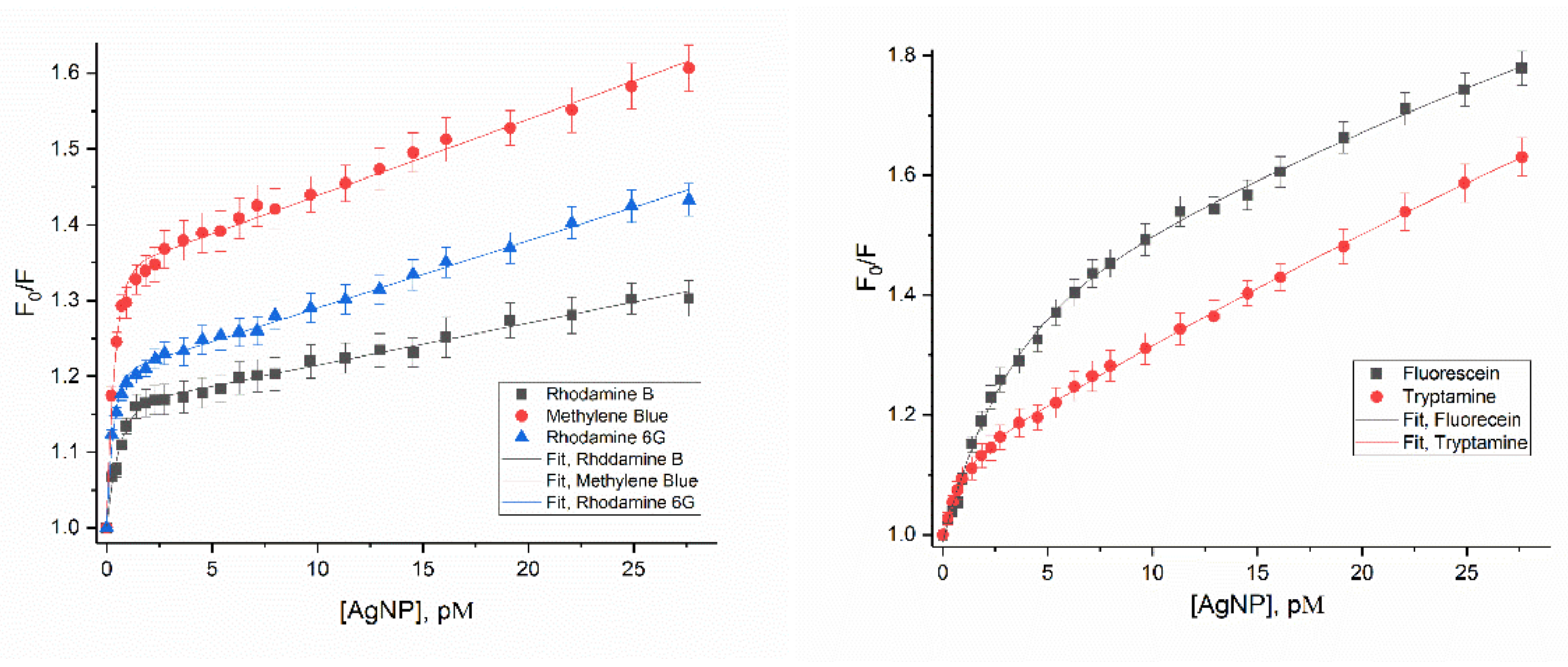
3.1. Effect of AgNPs on the Fluorescence of Dye Molecules
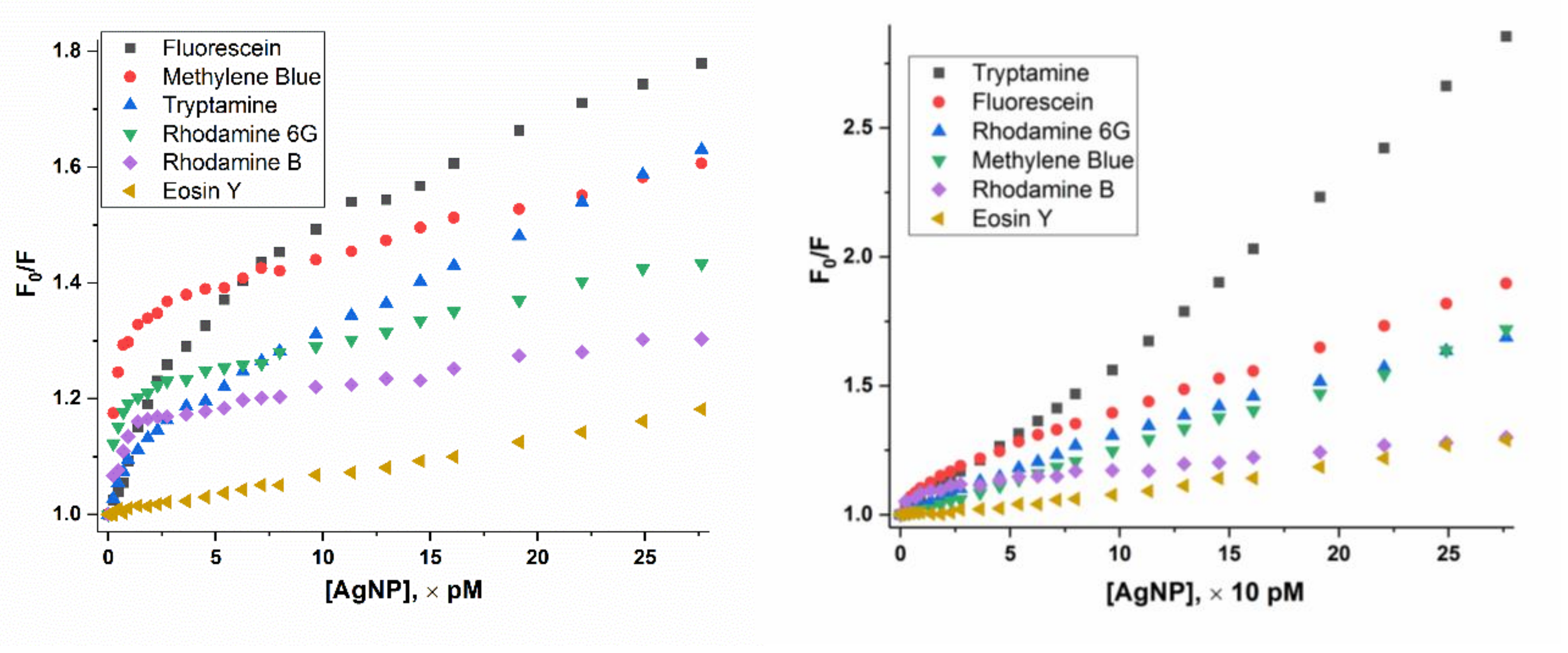
3.2. Different Behaviors of the Dyes on AgNP Surface
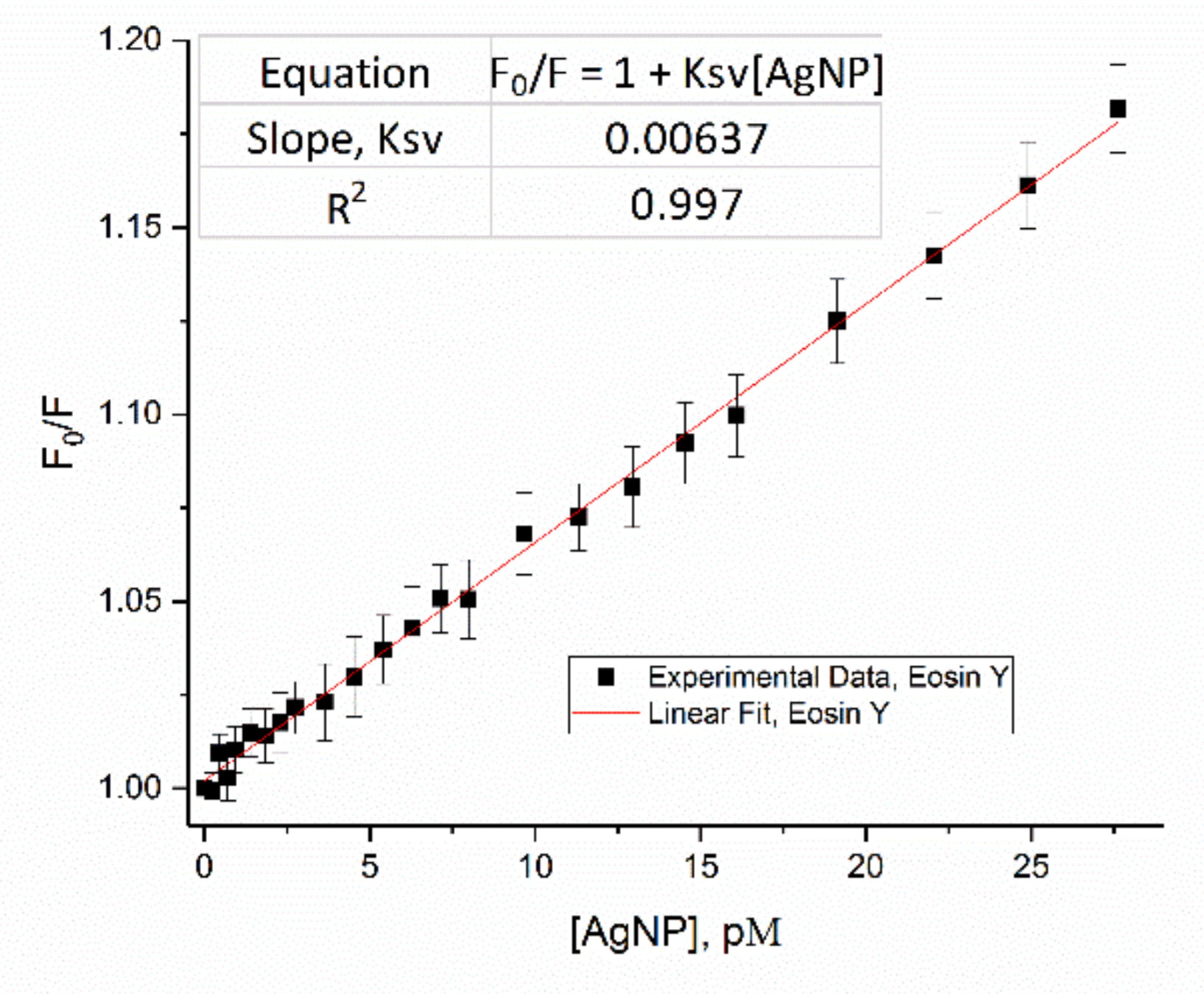
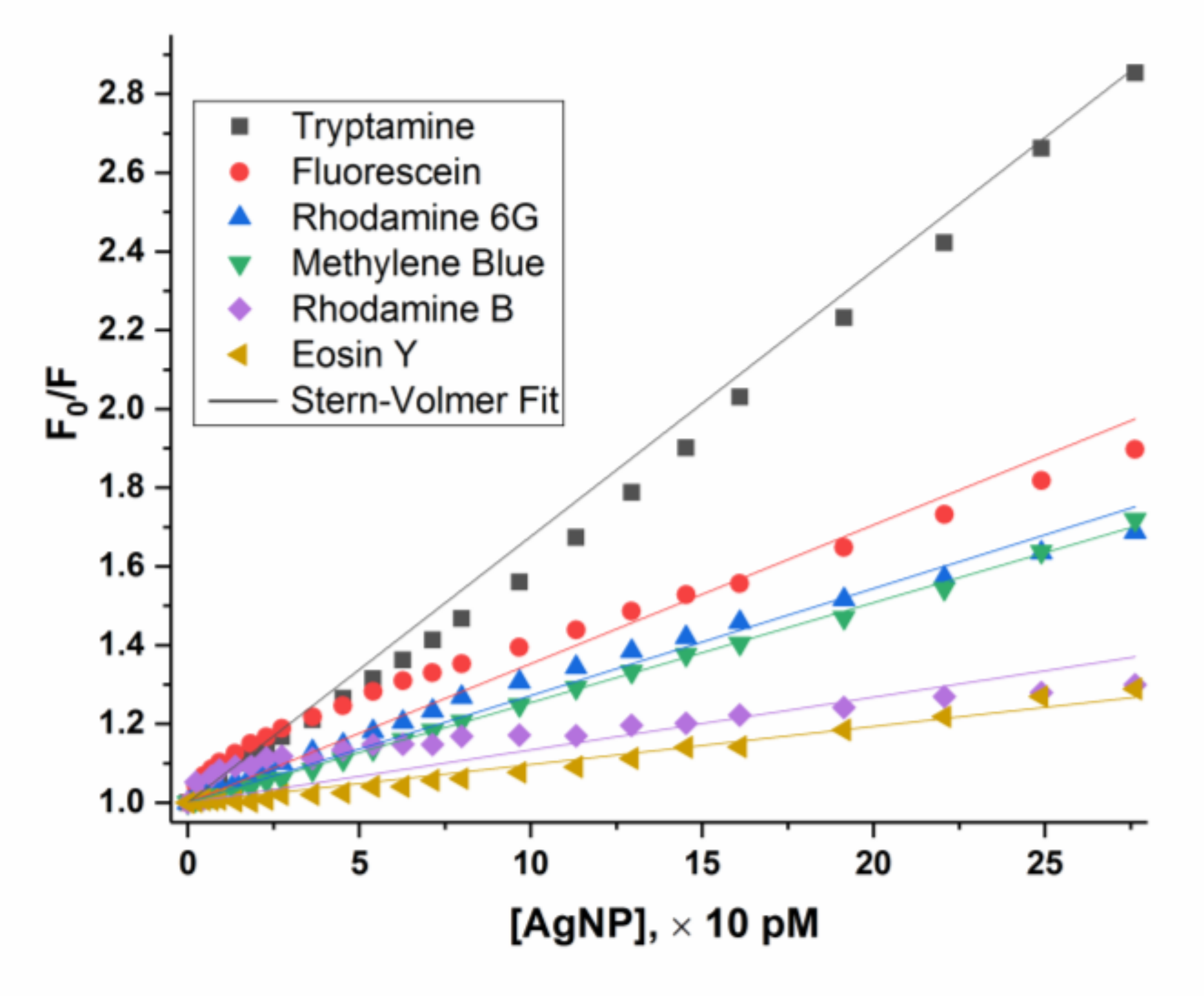
3.2.1. Stern-Volmer Fluorescence Quenching Model for eosin Y
3.2.2. Mathematics Model for the Molecular Self-Assembly on AgNP Surface
3.2.3. “Super” Stern-Volmer Fluorescence Quenching Constant
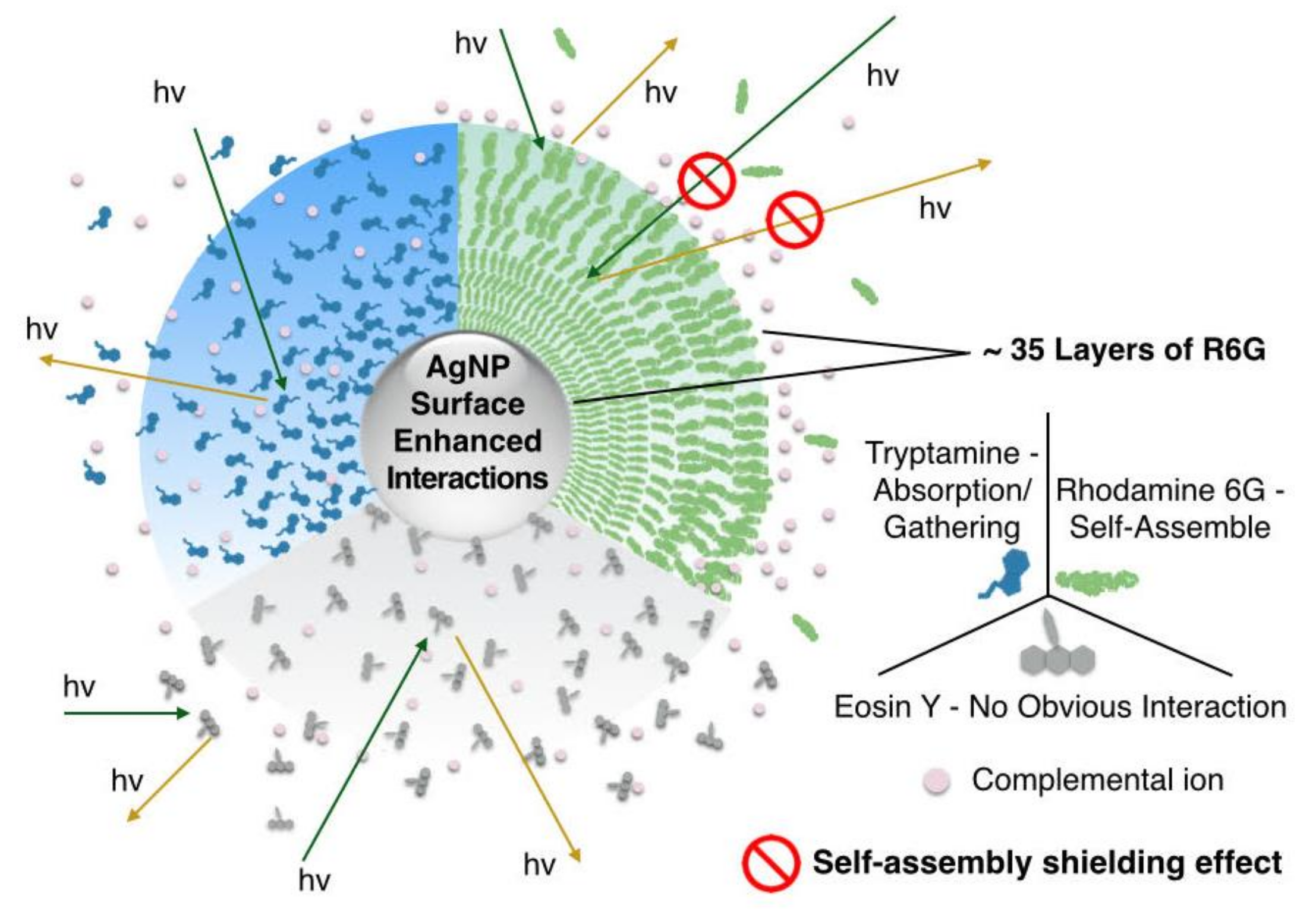
3.3. “Self-Assembly Shielding Effect” Caused Fluorescence Quenching
3.4. Factors Affecting Molecular Interactions on AgNP Interface
3.4.1. Concentration of both Dyes and Nanoparticles
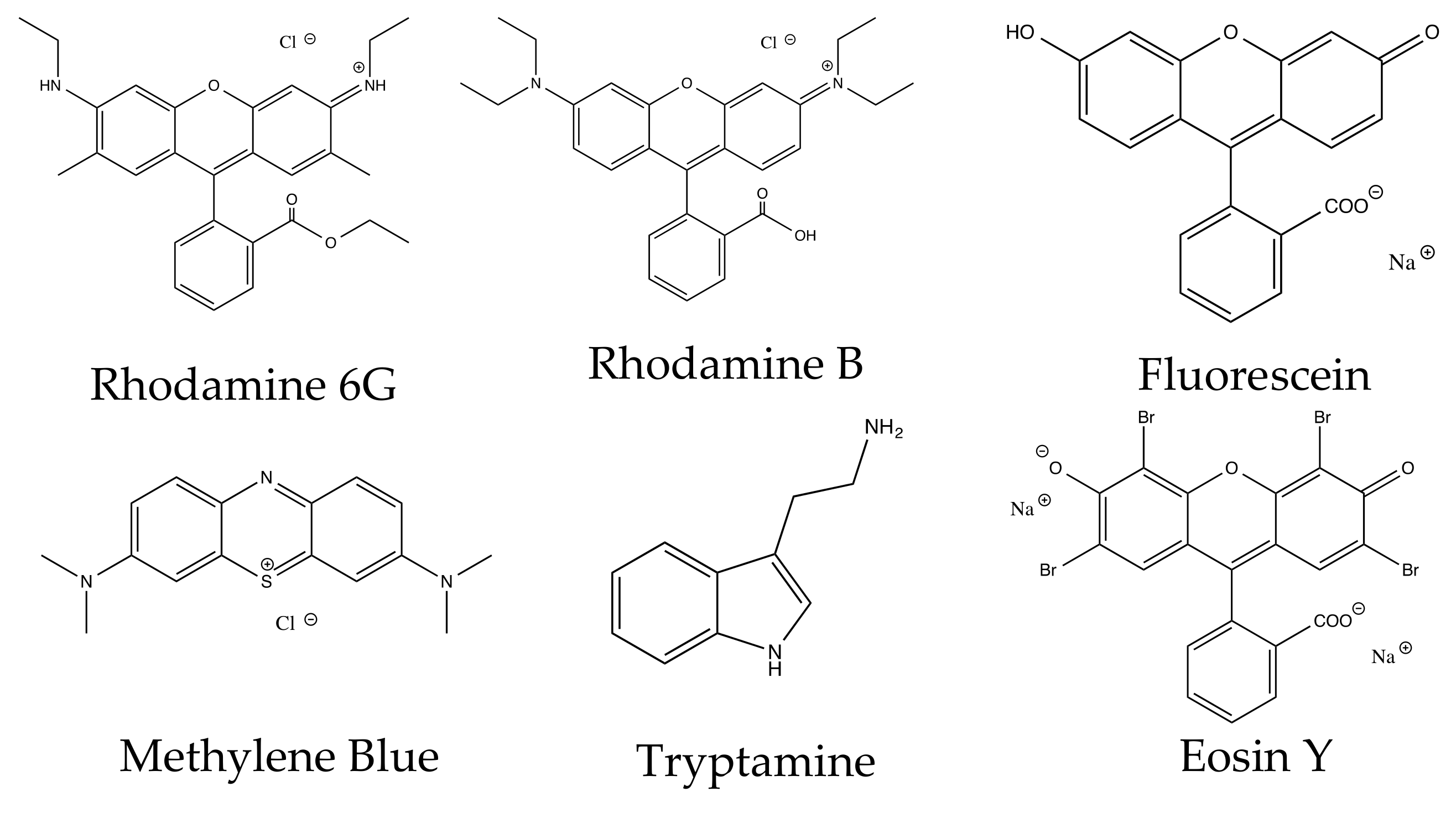
3.4.2. Intrinsic Molecular Structure
3.4.3. Charge
3.4.4. Functional Groups
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, W.; Lu, G.; Cui, C.; Liu, Y.; Li, S.; Yan, W.; Xing, C.; Chi, Y.R.; Yang, Y.; Huo, F. A Family of Metal-Organic Frameworks Exhibiting Size-Selective Catalysis with Encapsulated Noble-Metal Nanoparticles. Adv. Mater. 2014, 26, 4056–4060. [Google Scholar] [CrossRef] [PubMed]
- Howes, P.D.; Chandrawati, R.; Stevens, M.M. Colloidal nanoparticles as advanced biological sensors. Science 2014, 346, 1247390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.; Yoo, D.; Ling, D.; Cho, M.H.; Hyeon, T.; Cheon, J. Iron Oxide Based Nanoparticles for Multimodal Imaging and Magnetoresponsive Therapy. Chem. Rev. 2015, 115, 10637–10689. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Win, K.Y.; Liu, S.; Teng, C.P.; Zheng, Y.; Han, M.-Y. Surface-functionalized nanoparticles for biosensing and imaging-guided therapeutics. Nanoscale 2013, 5, 3127–3148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dasari, T.P.S.; Deng, H.; Yu, H. Antimicrobial Activity of Gold Nanoparticles and Ionic Gold. J. Environ. Sci. Health Part C 2015, 33, 286–327. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; McShan, D.; Zhang, Y.; Sinha, S.S.; Arslan, Z.; Ray, P.C.; Yu, H. Mechanistic study of the synergistic antibacterial activity of combined silver nanoparticles and common antibiotics. Environ. Sci. Technol. 2016, 50, 8840–8848. [Google Scholar] [CrossRef] [PubMed]
- Pourzahedi, L.; Eckelman, M.J. Environmental life cycle assessment of nanosilver-enabled bandages. Environ. Sci. Technol. 2014, 49, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, G.; Griffin, J.I.; Brenneman, B.; Banda, N.K.; Holers, V.M.; Backos, D.S.; Wu, L.; Moghimi, S.M.; Simberg, D. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotech. 2017, 12, 387–393. [Google Scholar] [CrossRef]
- Pulido-Reyes, G.; Leganes, F.; Fernández-Piñas, F.; Rosal, R. Bio-nano interface and environment: A critical review. Environ. Toxicol. Chem. 2017, 36, 3181–3193. [Google Scholar] [CrossRef]
- Deng, H.; Zhang, Y.; Yu, H. Nanoparticles considered as mixtures for toxicological research. J. Environ. Sci. Health Part C 2018, 36, 1–20. [Google Scholar] [CrossRef]
- Liu, C.; He, H.; Pandey, R.; Hussain, S.; Karna, S.P. Interaction of Metallic Nanoparticles with a Biologically Active Molecule, Dopamine. J. Phys. Chem. B 2008, 112, 15256–15259. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, Y.; Gao, J.; Zhang, Y.; Li, S. Interaction of Melamine Molecules with Silver Nanoparticles Explored by Surface-Enhanced Raman Scattering and Density Functional Theory Calculations. Appl. Spectrosc. 2013, 67, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Şenol, A.M.; Metin, Ö.; Acar, M.; Onganer, Y.; Meral, K. The interaction of fluorescent Pyronin Y molecules with monodisperse silver nanoparticles in chloroform. J. Mol. Struct. 2016, 1103, 212–216. [Google Scholar] [CrossRef]
- Choi, S.-H.; Lee, S.-H.; Hwang, Y.-M.; Lee, K.-P.; Kang, H.-D. Interaction between the surface of the silver nanoparticles prepared by γ-irradiation and organic molecules containing thiol group. Radiat. Phys. Chem. 2003, 67, 517–521. [Google Scholar] [CrossRef]
- Deng, H.; Yu, H. A mini review on controlling the size of Ag nanoclusters by changing the stabilizer to Ag ratio and by changing DNA sequence. Adv. Nat. Sci. 2015, 8, 1–9. [Google Scholar]
- Smith, D.S.; Bell, R.A.; Kramer, J.R. Metal speciation in natural waters with emphasis on reduced sulfur groups as strong metal binding sites. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2002, 133, 65–74. [Google Scholar] [CrossRef]
- Munro, C.H.; Smith, W.E.; Garner, M.; Clarkson, J.; White, P.C. Characterization of the Surface of a Citrate-Reduced Colloid Optimized for Use as a Substrate for Surface-Enhanced Resonance Raman Scattering. Langmuir 1995, 11, 3712–3720. [Google Scholar] [CrossRef]
- Petty, J.T.; Zheng, J.; Hud, N.V.; Dickson, R.M. DNA-Templated Ag Nanocluster Formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef]
- Zhao, J.; Jensen, L.; Sung, J.; Zou, S.; Schatz, G.C.; Van Duyne, R.P. Interaction of Plasmon and Molecular Resonances for Rhodamine 6G Adsorbed on Silver Nanoparticles. J. Am. Chem. Soc. 2007, 129, 7647–7656. [Google Scholar] [CrossRef]
- Basheer, N.S.; Kumar, B.R.; Kurian, A.; George, S.D. Thermal lens probing of distant dependent fluorescence quenching of Rhodamine 6G by silver nanoparticles. J. Lumin. 2013, 137, 225–229. [Google Scholar] [CrossRef]
- Tom, R.T.; Pradeep, T. Interaction of azide ion with hemin and cytochrome c immobilized on Au and Ag nanoparticles. Langmuir 2005, 21, 11896–11902. [Google Scholar] [CrossRef] [PubMed]
- Jadzinsky, P.D.; Calero, G.; Ackerson, C.J.; Bushnell, D.A.; Kornberg, R.D. Structure of a Thiol Monolayer-Protected Gold Nanoparticle at 1.1 A Resolution. Science 2007, 318, 430–433. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. A guide to simple and informative binding assays. Mol. Biol. Cell 2010, 21, 4061–4067. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.P.; Brown, J.S.; Bharti, B.; Wang, A.; Gangwal, S.; Houck, K.; Hubal, E.A.C.; Paunov, V.N.; Stoyanov, S.D.; Velev, O.D. An environmentally benign antimicrobial nanoparticle based on a silver-infused lignin core. Nat. Nanotechnol. 2015, 10, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Tassa, C.; Duffner, J.L.; Lewis, T.A.; Weissleder, R.; Schreiber, S.L.; Koehler, A.N.; Shaw, S.Y. Binding affinity and kinetic analysis of targeted small molecule-modified nanoparticles. Bioconjugate Chem. 2009, 21, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Shewale, V.; Pandey, R.; Shanker, V.; Hussain, S.; Karna, S.P. Tryptophan–Gold Nanoparticle Interaction: A First-Principles Quantum Mechanical Study. J. Phys. Chem. C 2011, 115, 22818–22826. [Google Scholar] [CrossRef]
- Deng, H.; Yu, H. Self-assembly of rhodamine 6G on silver nanoparticles. Chem. Phys. Lett. 2018, 692, 75–80. [Google Scholar] [CrossRef]
- Lacerda, S.H.D.P.; Park, J.J.; Meuse, C.; Pristinski, D.; Becker, M.L.; Karim, A.; Douglas, J.F. Interaction of Gold Nanoparticles with Common Human Blood Proteins. ACS Nano 2009, 4, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis; John Wiley & Sons: New York, NY, USA, 2004. [Google Scholar]
- Deng, H.; Gao, Y.; Dasari, T.P.S.; Ray, P.C.; Yu, H. A facile 3D construct of graphene oxide embedded with silver nanoparticles and its potential application as water filter. J. Miss. Acad. Sci. 2016, 61, 190–197. [Google Scholar]
- Tejamaya, M.; Römer, I.; Merrifield, R.C.; Lead, J.R. Stability of citrate, PVP, and PEG coated silver nanoparticles in ecotoxicology media. Environ. Sci. Technol. 2012, 46, 7011–7017. [Google Scholar] [CrossRef]
- Sun, Y.; Xia, Y. Shape-Controlled Synthesis of Gold and Silver Nanoparticles. Science 2002, 298, 2176–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Atwater, M.; Wang, J.; Huo, Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf. B Biointerfaces 2007, 58, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Henglein, A.; Giersig, M. Formation of Colloidal Silver Nanoparticles: Capping Action of Citrate. J. Phys. Chem. B 1999, 103, 9533–9539. [Google Scholar] [CrossRef]
- Santhi, A.; Umadevi, M.; Ramakrishnan, V.; Radhakrishnan, P.; Nampoori, V. Effect of silver nano particles on the fluorescence quantum yield of Rhodamine 6G determined using dual beam thermal lens method. Spectrochim. Acta A 2004, 60, 1077–1083. [Google Scholar] [CrossRef]
- Chakraborty, M.; Panda, A.K. Spectral behaviour of eosin Y in different solvents and aqueous surfactant media. Spectrochim. Acta A 2011, 81, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, Z.; Schwille, P. Precise measurement of diffusion coefficients using scanning fluorescence correlation spectroscopy. Biophys. J. 2008, 94, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching of indole and model micelle systems. J. Phys. Chem. 1976, 80, 486–493. [Google Scholar] [CrossRef]
- Ghosh, D.; Chattopadhyay, N. Gold and silver nanoparticles based superquenching of fluorescence: A review. J. Lumin. 2015, 160, 223–232. [Google Scholar] [CrossRef]
- Long, Z.; Liu, M.; Mao, L.; Zeng, G.; Huang, Q.; Huang, H.; Deng, F.; Wan, Y.; Zhang, X.; Wei, Y. One-step synthesis, self-assembly and bioimaging applications of adenosine triphosphate containing amphiphilies with aggregation-induced emission feature. Mater. Sci. Eng. C 2017, 73, 252–256. [Google Scholar] [CrossRef]
- Quinn, S.D.; Dalgarno, P.A.; Cameron, R.T.; Hedley, G.J.; Hacker, C.; Lucocq, J.M.; Baillie, G.S.; Samuel, I.D.W.; Penedo, J.C. Real-time probing of β-amyloid self-assembly and inhibition using fluorescence self-quenching between neighbouring dyes. Mol. BioSyst. 2014, 10, 34–44. [Google Scholar] [CrossRef]
- Lübtow, M.; Helmers, I.; Stepanenko, V.; Albuquerque, R.Q.; Marder, T.B.; Fernández, G. Self-Assembly of 9,10-Bis(phenylethynyl) Anthracene (BPEA) Derivatives: Influence of π-π and Hydrogen-Bonding Interactions on Aggregate Morphology and Self-Assembly Mechanism. Chem. A Eur. J. 2017, 23, 6198–6205. [Google Scholar] [CrossRef] [PubMed]
- Nador, F.; Wnuk, K.; Roscini, C.; Solorzano, R.; Faraudo, J.; Ruiz-Molina, D.; Novio, F. Solvent-Tuned Supramolecular Assembly of Fluorescent Catechol/Pyrene Amphiphilic Molecules. Chem. A Eur. J. 2018, 24, 14724–14732. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.-C.; Mata, J.P.; Wu, C.-M.; Lo, C.-T. Morphology-Mediated Photoresponsive and Fluorescence Behaviors of Azobenzene-Containing Block Copolymers. Langmuir 2018, 34, 7416–7427. [Google Scholar] [CrossRef] [PubMed]
- Lynch, I.; Salvati, A.; Dawson, K.A. Protein-nanoparticle interactions: What does the cell see? Nat. Nanotech. 2009, 4, 546–547. [Google Scholar] [CrossRef] [PubMed]
- Al-Thabaiti, N.S.; Malik, M.A.; Khan, Z. Protein interactions with silver nanoparticles: Green synthesis, and biophysical approach. Int. J. Biol. Macromol. 2017, 95, 421–428. [Google Scholar] [CrossRef] [PubMed]
| Dye | Excitation Wavelength, nm | Slit Width, nm, 1 µM | Slit Width, nm, 10 µM | Emission Peak, nm |
|---|---|---|---|---|
| Rhodamine 6G | 500 | 1 | 1 | 554 |
| Rhodamine B | 540 | 2 | 1 | 580 |
| Methylene Blue | 640 | 5 | 4 | 688 |
| Fluorescein | 470 | 1 | 1 | 513 |
| Eosin Y | 505 | 2 | 1 | 539 |
| Tryptamine | 280 | 2 | 2 | 356 |
| AgNP with citrate | Rhodamine 6G | Rhodamine B | Methylene Blue * | Fluorescein | Eosin Y | Tryptamine | |
|---|---|---|---|---|---|---|---|
| Size, nm | 33.4 ± 1.6 | 318.5 ± 34.4 | 197.6 ± 62.1 | / | 67.1 ± 12.4 | 42.8 ± 3.9 | 69.2 ± 13.0 |
| Potential, mV | −47.3 ± 1.2 | 21.5 ± 2.0 | 17.6 ± 1.8 | / | −16.9 ± 2.2 | −31.8 ± 1.6 | 8.9 ± 3.7 |
| Dye | A | B | Ksv, nM−1 (AgNP) | Ksv, M−1 (Ag) | R2 | Dyes/AgNP | |
|---|---|---|---|---|---|---|---|
| Rhodamine B | 0.16 | 0.59 | 5.5 | 13,000 | 0.992 | 180,000 | |
| Rhodamine 6G | 0.20 | 0.33 | 8.9 | 21,000 | 0.996 | 220,000 | |
| Methylene Blue | 0.33 | 0.38 | 10 | 24,000 | 0.995 | 200,000 | |
| Fluorescein | 0.37 | 3.37 | 1.5 | 36,000 | 0.998 | / | |
| Tryptamine | 0.12 | 1.27 | 1.8 | 43,000 | 0.999 | / | |
| Eosin Y | / | / | 6.4 | 15,000 | 0.997 | / | |
| Tryptamine* | / | / | / | 33* | / | / | |
| Tryptamine | Fluorescein | R6G | Methylene Blue | Rhodamine B | Eosin Y | |
|---|---|---|---|---|---|---|
| Ksv | 68 | 35 | 27 | 25 | 13 | 10 |
| Dye | Charge | Functional Groups | Pair Ions | Self-Assembly |
|---|---|---|---|---|
| Rhodamine 6G | 1+ | -NH-, -N=, -O-, -COO- | Cl- | Yes |
| Rhodamine B | 1+ | -N-, -N=, -O-, -COO- | Cl- | Yes |
| Methylene Blue | 1+ | -N-, -N=, -S- | Cl- | Yes |
| Fluorescein | 1- | -OH, -COO-, -O-, =O | Na+ | Weak |
| Eosin Y | 2- | -COO-, –Br, -O-, =O | Na+ | No |
| Tryptamine | 0 | -NH-, -NH2 | NA | Weak |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, H.; Yu, H. Silver Nanoparticle Surface Enabled Self-Assembly of Organic Dye Molecules. Materials 2019, 12, 2592. https://doi.org/10.3390/ma12162592
Deng H, Yu H. Silver Nanoparticle Surface Enabled Self-Assembly of Organic Dye Molecules. Materials. 2019; 12(16):2592. https://doi.org/10.3390/ma12162592
Chicago/Turabian StyleDeng, Hua, and Hongtao Yu. 2019. "Silver Nanoparticle Surface Enabled Self-Assembly of Organic Dye Molecules" Materials 12, no. 16: 2592. https://doi.org/10.3390/ma12162592