Preliminary Molecular Dynamic Simulations of the Estrogen Receptor Alpha Ligand Binding Domain from Antagonist to Apo
Abstract
:Introduction
Methods
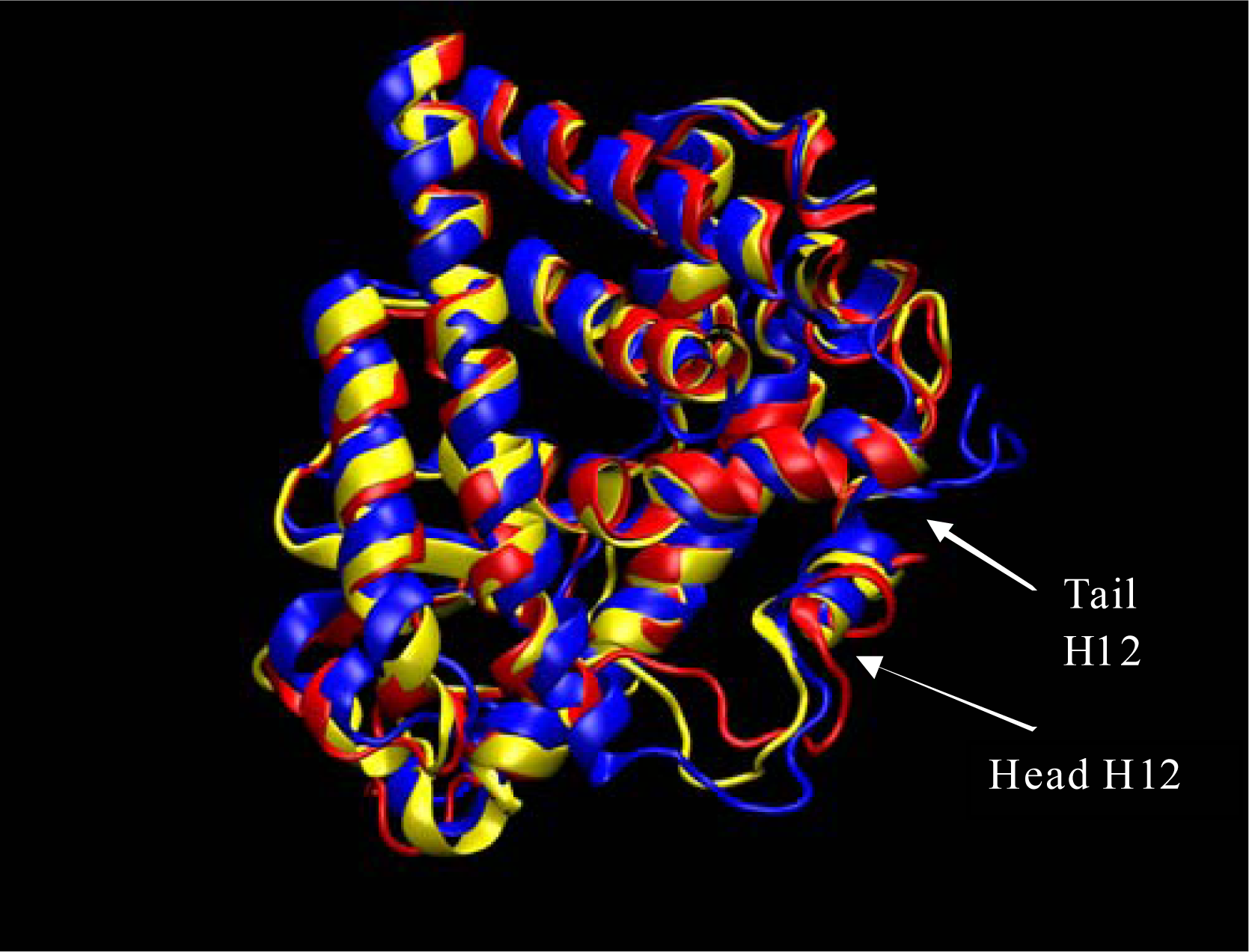
Results
Conclusions
Acknowledgments
References
- Sumbayev, VV; Bonefeld-Jørgensen, EC; Wind, T; Andreasen, PA. A novel pesticide-induced conformational state of the oestrogen receptor ligand-binding domain, detected by conformation-specific peptide binding. FEBS Lett 2005, 579, 541–548. [Google Scholar]
- Shiau, AK; Barstad, D; Loria, PM; Cheng, L; Kushner, PJ; Agard, DA; Greene, GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 1998, 95, 927–937. [Google Scholar]
- Tanenbaum, DM; Wang, Y; Williams, SP; Sigler, PB. Crystallographic comparison of the estrogen and pogesterone receptor’s ligand binding domain. Proc Nat Acad Sci USA 1998, 95, 5998–6003. [Google Scholar]
- Laak, AM; Jongejan, A; Vermeulen, NP; Wamelink, M; Geerke, D; Meerman, JH. Prediction of ligand binding affinity and orientation of xenoestrogens to the estrogen receptor by molecular dynamics simulations and the linear interaction energy method. J Med Chem 2004, 47(4), 1018–30. [Google Scholar]
- Nilsson, S; Makela, S; Treuter, E; Tujague, M; Thomesen, J; Andersson, G; Enmark, E; Pettersson, K; Warner, M; Gustafsson, JA. Mechanisms of Estrogne action. Physiol Rev 2001, 81, 1535–1565. [Google Scholar]
- McKenna, NM; O’Malley, BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 2002, 108, 465–474. [Google Scholar]
- Celik, L; Lund, JDD; Schiøtt, B. Conformational dynamics of the Estrogen Receptor a: Moleulcar Dynamics Simulations of the Influence of Binding Site Structure on Protein Dynamics. Biochemistry 2007, 46, 1743–1758. [Google Scholar]
- Case, DA; Cheatham, TE, III; Darden, T; Gohlke, H; Luo, R; Merz, KM, Jr; Onufriev, A; Simmerling, C; Wang, B; et al. The Amber biomolecular simulation programs. J Comput Chem 2005, 26, 1668–1688. [Google Scholar]
- Pearlman, DA; Case, DA; Caldwell, JW; Ross, WR; Cheatham, TE, III; DeBolt, S; Ferguson, D; Seibel, G; Kollman, P. AMBER, a computer program for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to elucidate the structures and energies of molecules. Comp Phys Commun 1995, 91, 1–41. [Google Scholar]
- Tuckerman, ME; Matyna, GJ. Understanding modern molecular dynamics: Techniques and applications. J Phys Chem B 2000, 104, 159–178. [Google Scholar]
- Martyna, GJ; Tobias, DJ; Klein, ML. Constant-pressure molecular dynamic algorithms. J Chem Phys 1994, 101, 4177–4189. [Google Scholar]
- Berendsen, HJC; Postma, JPM; van Gunsteren, WF; Dinola, A; Haak, JR. Molecular dynamics with coupling to an external bath. J Chem Phys 1984, 81, 3684–3690. [Google Scholar]
- Humphrey, W; Dalke, A; Schulten, K. VMD - visual molecular dynamics. J Molec Graphics 1996, 14, 33–38. [Google Scholar]


© MDPI. All rights reserved
Share and Cite
McGee, T.D.; Edwards, J.; Roitberg, A.E. Preliminary Molecular Dynamic Simulations of the Estrogen Receptor Alpha Ligand Binding Domain from Antagonist to Apo. Int. J. Environ. Res. Public Health 2008, 5, 111-114. https://doi.org/10.3390/ijerph5020111
McGee TD, Edwards J, Roitberg AE. Preliminary Molecular Dynamic Simulations of the Estrogen Receptor Alpha Ligand Binding Domain from Antagonist to Apo. International Journal of Environmental Research and Public Health. 2008; 5(2):111-114. https://doi.org/10.3390/ijerph5020111
Chicago/Turabian StyleMcGee, T. Dwight, Jesse Edwards, and Adrian E. Roitberg. 2008. "Preliminary Molecular Dynamic Simulations of the Estrogen Receptor Alpha Ligand Binding Domain from Antagonist to Apo" International Journal of Environmental Research and Public Health 5, no. 2: 111-114. https://doi.org/10.3390/ijerph5020111



