The Screening and Identification of a Dextranase-Secreting Marine Actinmycete Saccharomonospora sp. K1 and Study of Its Enzymatic Characteristics
Abstract
:1. Introduction
2. Results
2.1. Screening and Identification of Strains
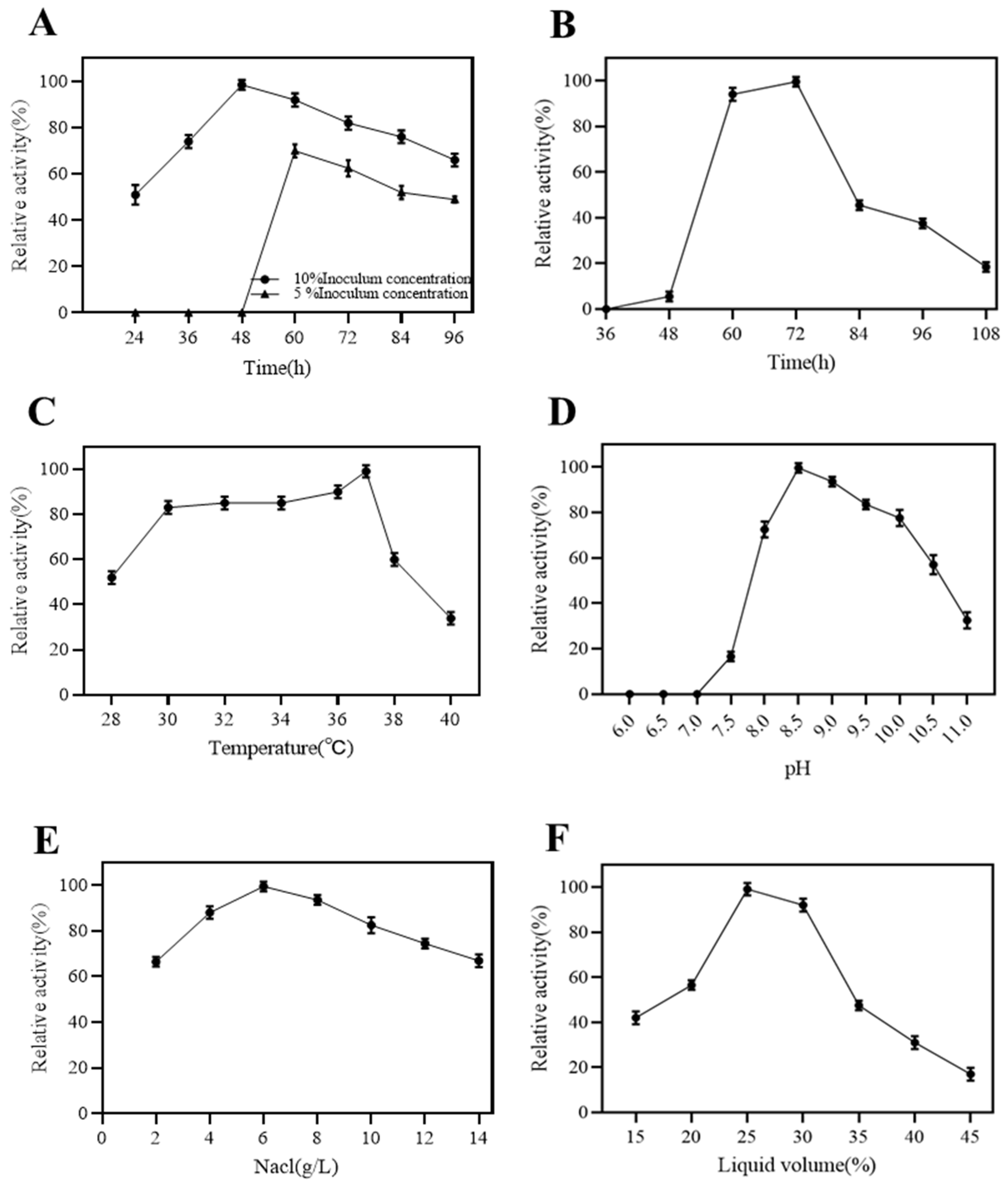
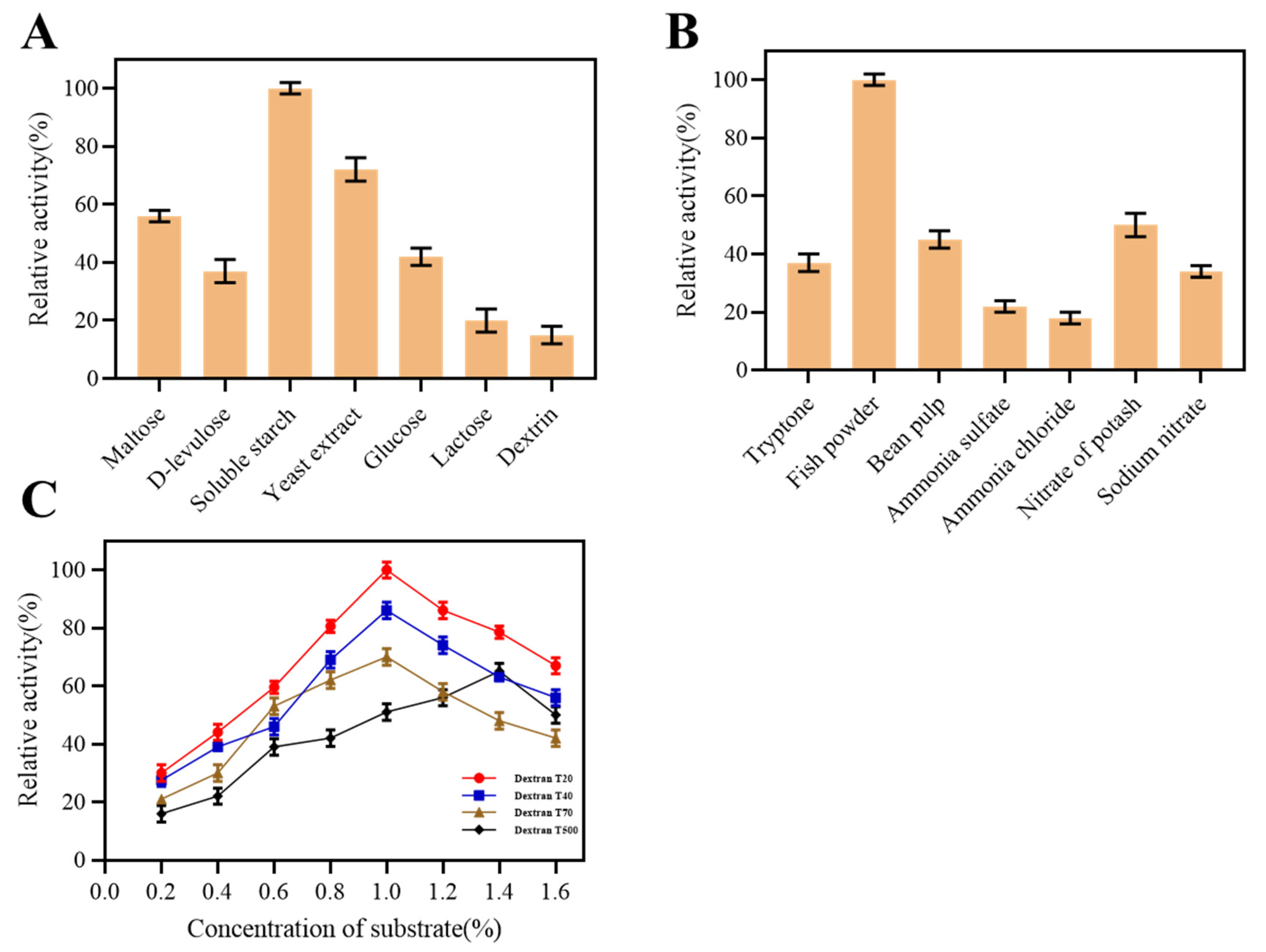
2.2. Fermentation Condition of Dextranase
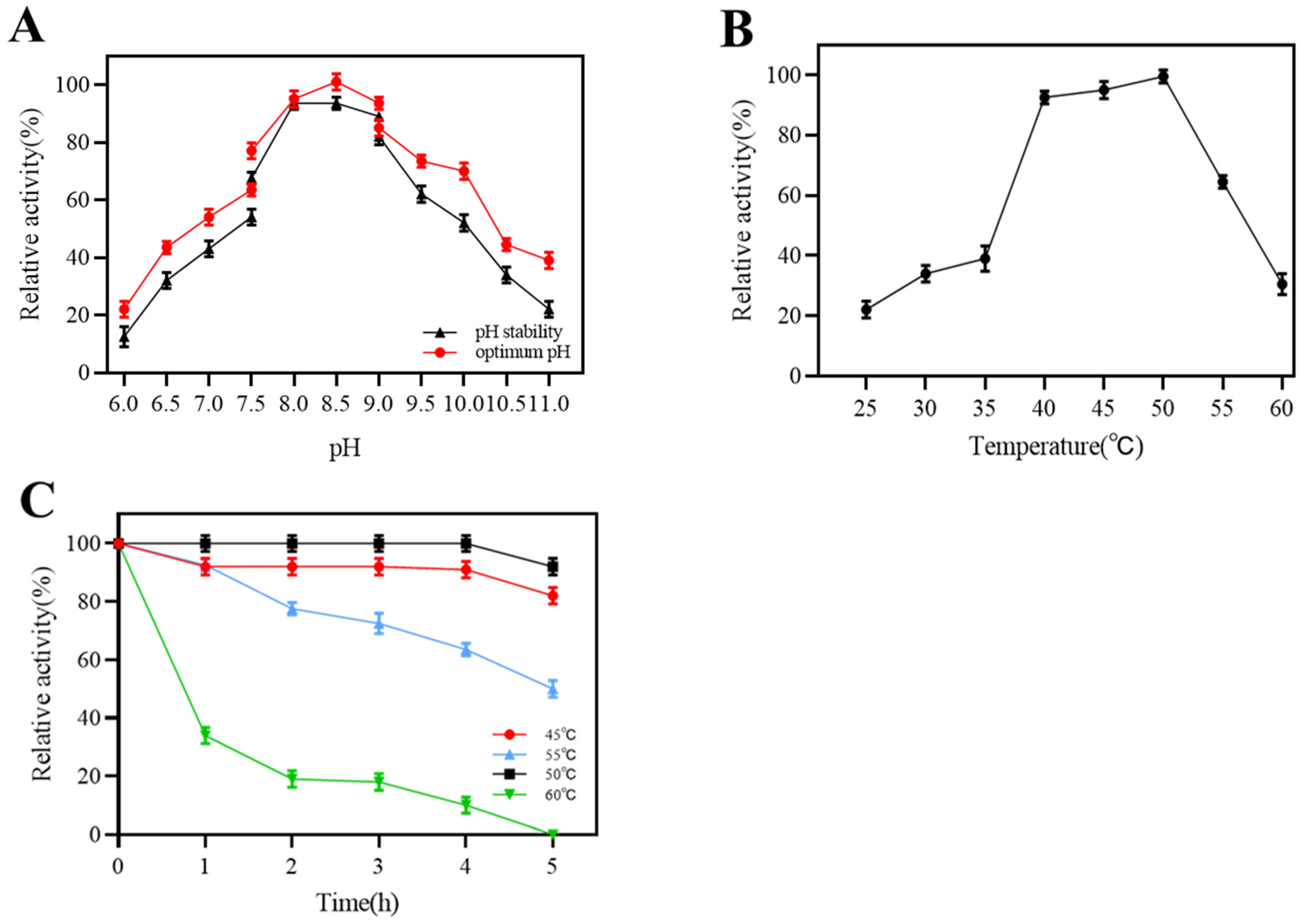
2.3. Characteristics of Dextranase
2.3.1. Effect of Temperature and pH on the Activity of Dextranase
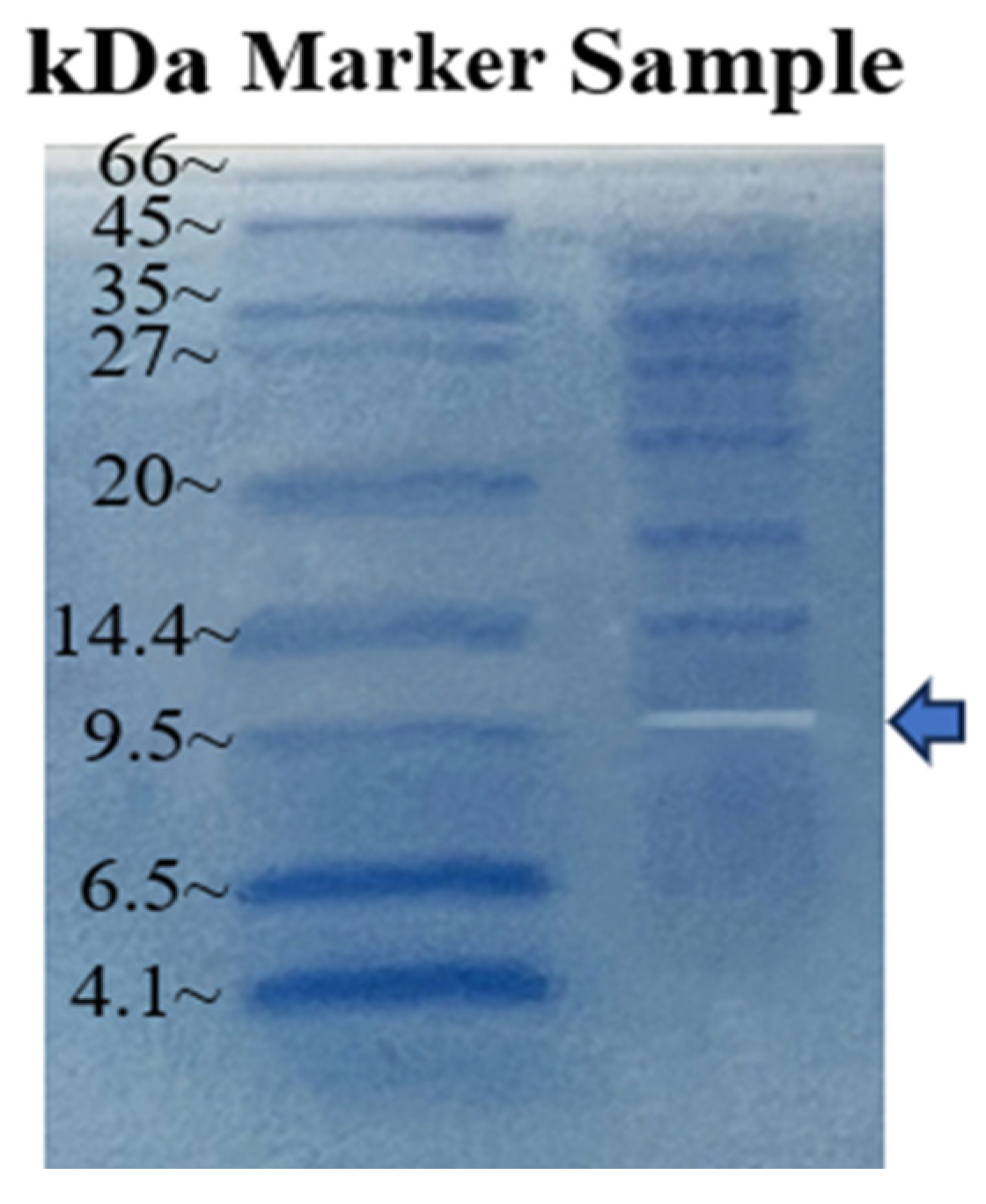
2.3.2. Molecular Weight of Dextranase
2.3.3. Effect of Metal Ions and Substrate Specificity
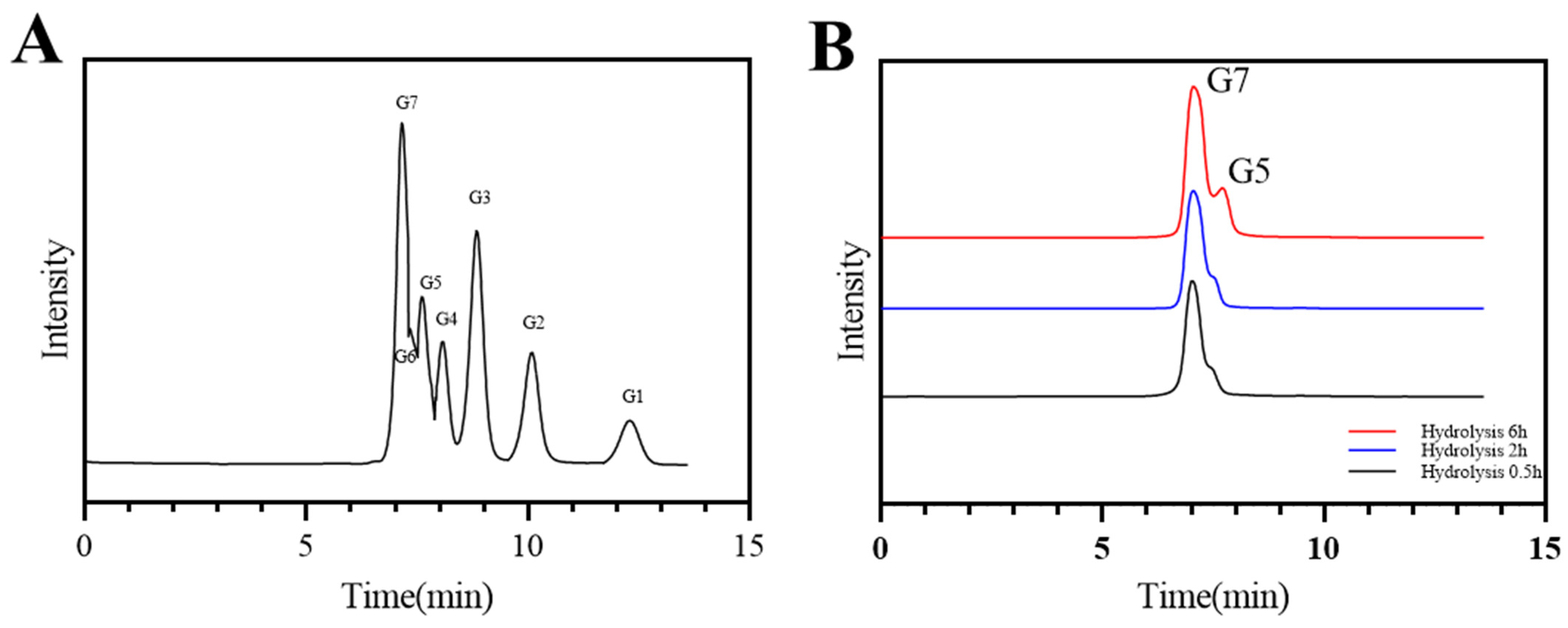
2.3.4. Analysis of Hydrolysates
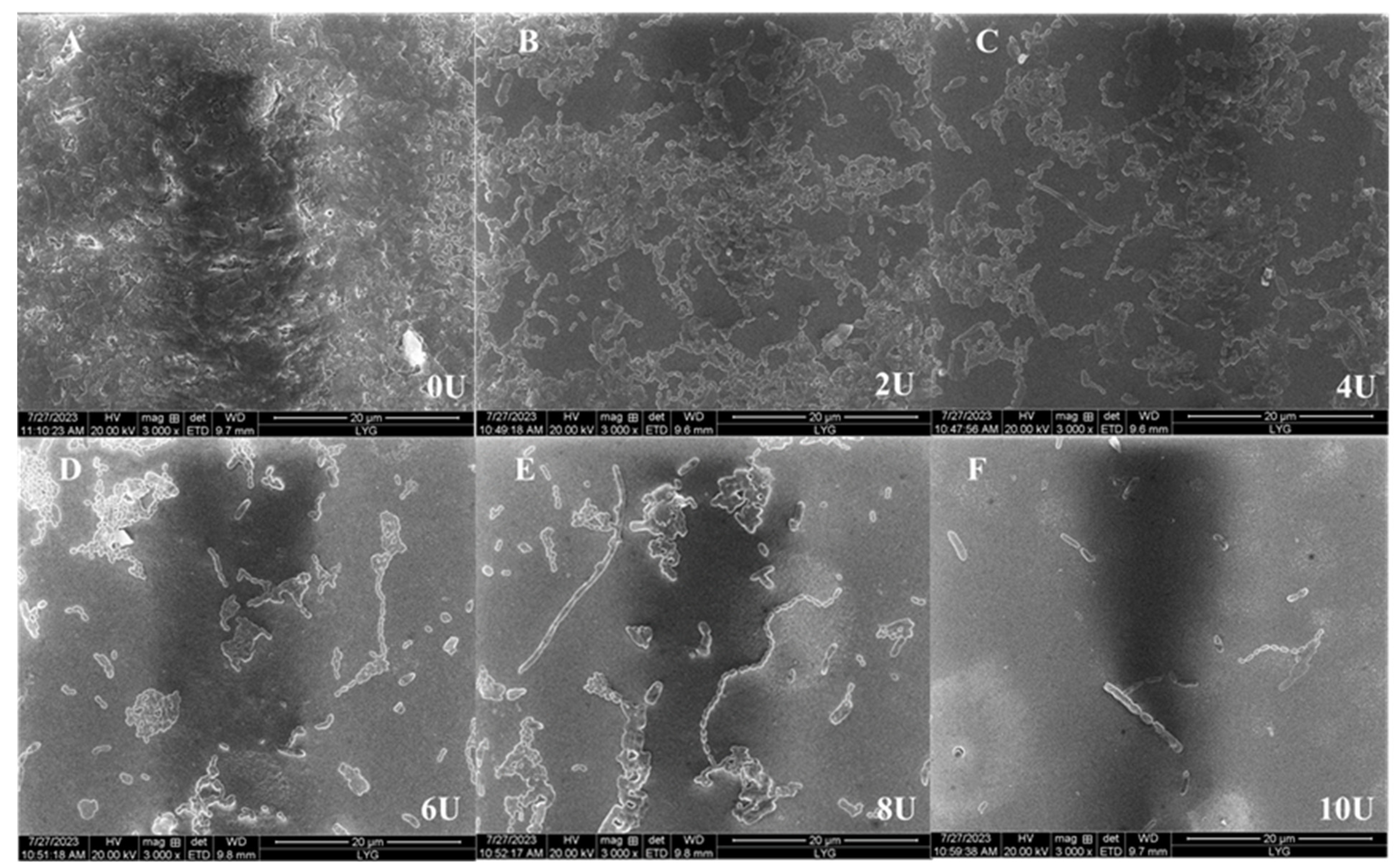
2.3.5. Effects of Dextranase on Biofilm
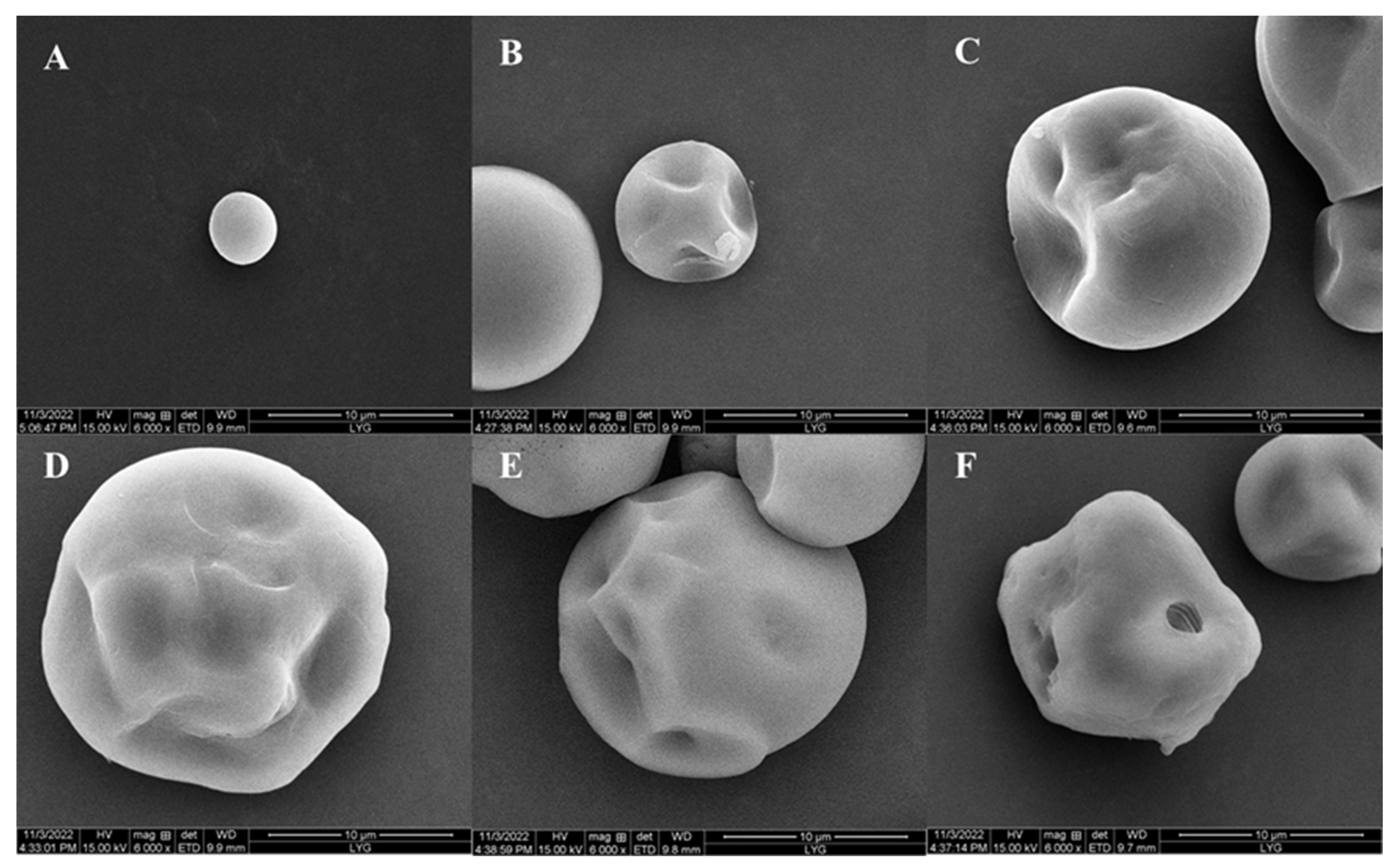
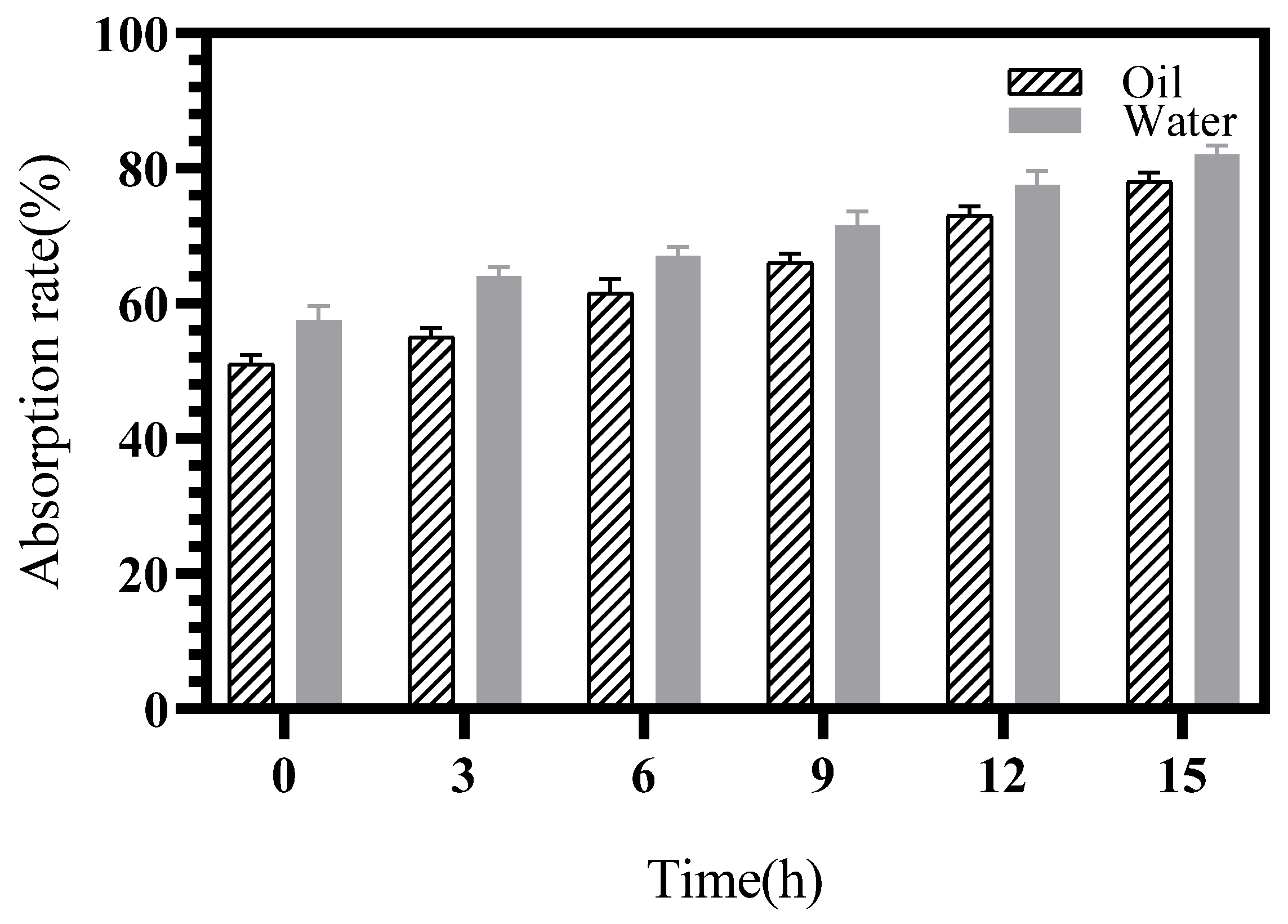
2.3.6. Porous Starch of Sweet Potato
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Screening of Dextranase-Producing Strains
4.2.2. Identification of Strain K1
4.2.3. Fermentation of Dextranase
4.2.4. Conditions for Dextranase Production
4.2.5. Enzymatic Properties of Dextranase
4.2.6. Analysis of Hydrolysates
4.2.7. Biofilm Removal by Dextranase
4.2.8. Preparation of Porous Starch and Determination of Water and Oil Absorption
4.2.9. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bhatia, S.; Bhakri, G.; Arora, M.; Uppal, S.K.; Batta, S.K. Dextranase production from paecilomyces lilacinus and its application for dextran removal from sugarcane juice. Sugar Tech 2010, 12, 133–138. [Google Scholar] [CrossRef]
- Khalikova, E.; Susi, P.; Korpela, T. Microbial dextran-hydrolyzing enzymes: Fundamentals and applications. Microbiol. Mol. Biol. Rev. 2005, 69, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Pittrof, S.L.; Kaufhold, L.; Fischer, A.; Wefers, D. Products released from structurally different dextrans by bacterial and fungal dextranases. Foods 2021, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Thitaram, S.N.; Chung, C.-H.; Day, D.F.; Hinton, A., Jr.; Bailey, J.S.; Siragusa, G.R. Isomaltooligosaccharide increases cecal bifidobacterium population in young broiler chickens. Poult. Sci. 2005, 84, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, F.A.; Jördening, H.-J. Immobilization of dextranase from Chaetomium erraticum. J. Biotechnol. 2007, 131, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Bowen, W.H.; Koo, H. Biology of streptococcus mutans-derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011, 45, 69–86. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Food Contact Materials; Enzymes and Processing Aids (CEP); Lambré, C.; Barat Baviera, J.M.; Bolognesi, C.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E. Safety evaluation of the food enzyme dextranase from the Collariella gracilis strain ae-dx. EFSA J. 2022, 20, e07279. [Google Scholar] [CrossRef]
- Falconer, D.J.; Mukerjea, R.; Robyt, J.F. Biosynthesis of dextrans with different molecular weights by selecting the concentration of leuconostoc mesenteroides b-512fmc dextransucrase, the sucrose concentration, and the temperature. Carbohydr. Res. 2011, 346, 280–284. [Google Scholar] [CrossRef]
- Robyt, J.F.; Yoon, S.-H.; Mukerjea, R. Dextransucrase and the mechanism for dextran biosynthesis. Carbohydr. Res. 2008, 343, 3039–3048. [Google Scholar] [CrossRef]
- Bu, Q.-T.; Li, Y.-P.; Xie, H.; Li, J.F.; Lv, Z.Y.; Su, Y.T.; Li, Y.Q. Rational engineering strategies for achieving high-yield, high-quality and high-stability of natural product production in actinomycetes. Metab. Eng. 2021, 67, 198–215. [Google Scholar] [CrossRef]
- de Borba Gurpilhares, D.; Cinelli, L.P.; Simas, N.K.; Pessoa, A., Jr.; Sette, L.D. Marine prebiotics: Polysaccharides and oligosaccharides obtained by using microbial enzymes. Food Chem. 2019, 280, 175–186. [Google Scholar] [CrossRef]
- Kim, S.; Le, T.C.; Han, S.-A.; Hillman, P.F.; Hong, A.; Hwang, S.; Du, Y.E.; Kim, H.; Oh, D.C.; Cha, S.S.; et al. Saccharobisindole, neoasterric methyl ester, and 7-chloro-4(1h)-quinolone: Three new compounds isolated from the marine bacterium Saccharomonospora sp. Mar. Drugs 2021, 20, 35. [Google Scholar] [CrossRef]
- Purushe, S.; Prakash, D.; Nawani, N.N.; Dhakephalkar, P.; Kapadnis, B. Biocatalytic potential of an alkalophilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manu-facture. Bioresour. Technol. 2012, 115, 2–7. [Google Scholar] [CrossRef]
- Lenkkeri, A.M.; Pienihäkkinen, K.; Hurme, S.; Alanen, P. The caries-preventive effect of xylitol/maltitol and erythritol/maltitol lozenges: Results of a double-blinded, cluster-randomized clinical trial in an area of natural fluoridation. Int. J. Paediatr. Dent. 2011, 22, 180–190. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Liu, B.-L.; Zheng, X.-H.; Huang, X.J.; Li, H.Y.; Zhang, Y.; Zhang, T.T.; Sun, D.Y.; Lin, B.R.; Zhou, G.X. Anandins a and b, two rare steroidal alkaloids from a marine Streptomyces anandii H41-59. Mar. Drugs 2017, 15, 355. [Google Scholar] [CrossRef] [PubMed]
- Arruda, A.O.; Senthamarai Kannan, R.; Inglehart, M.R.; Rezende, C.T.; Sohn, W. Effect of 5% fluoride varnish application on caries among school children in rural brazil: A randomized controlled trial. Community Dent. Oral Epidemiol. 2011, 40, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Rajan, N.; Biswas, P.; Banerjee, R. A novel approach for resistant starch production from green banana flour using amylopullulanase. LWT-Food Sci. Technol. 2022, 153, 112391. [Google Scholar] [CrossRef]
- Fuentes-Zaragoza, E.; Riquelme-Navarrete, M.J.; Sánchez-Zapata, E.; Pérez-Álvarez, J.A. Resistant starch as functional ingredient: A review. Food Res. Int. 2010, 43, 931–942. [Google Scholar] [CrossRef]
- Zhang, Y.-Q.; Li, R.-H.; Zhang, H.-B.; Wu, M.; Hu, X.Q. Purification, characterization, and application of a thermostable dextranase from Talaromyces pinophilus. J. Ind. Microbiol. Biotechnol. 2016, 44, 317–327. [Google Scholar] [CrossRef]
- Wynter, C.V.A.; Chang, M.; De Jersey, J.; Patel, B.; Inkerman, P.A.; Hamilton, S. Isolation and characterization of a thermostable dextranase. Enzym. Microb. Technol. 1997, 20, 242–247. [Google Scholar] [CrossRef]
- Koenig, D.; Day, D. The purification and characterization of a dextranase from Lipomyces starkeyi. Eur. J. Biochem. 1989, 183, 161–167. [Google Scholar] [CrossRef]
- Eggleston, G.; Dilks, A.; Blowers, M.; Winters, K. Successful application of dextranase in sugar beet factories. In Proceedings of the 36 th Biennial Meeting of the American Society of Sugar Beet Technologists, Albuquerque, NM, USA, 2–5 March 2011. [Google Scholar]
- Deng, T.; Feng, Y.; Xu, L.; Tian, X.; Lai, X.; Lyu, M.; Wang, S. Expression, purification and characterization of a cold-adapted dextranase from marine bacteria and its ability to remove dental plaque. Protein Expr. Purif. 2020, 174, 105678. [Google Scholar] [CrossRef]
- Wu, D.-T.; Zhang, H.-B.; Huang, L.-J.; Hu, X.Q. Purification and characterization of extracellular dextranase from a novel producer, hypocrea lixii f1002, and its use in oligodextran production. Process Biochem. 2011, 46, 1942–1950. [Google Scholar] [CrossRef]
- Lai, X.; Liu, X.; Liu, X.; Deng, T.; Feng, Y.; Tian, X.; Lyu, M.; Wang, S. The marine Catenovulum agarivorans mnh15 and dextranase: Removing dental plaque. Mar. Drugs 2019, 17, 592. [Google Scholar] [CrossRef]
- Zohra, R.R.; Aman, A.; Ansari, A.; Haider, M.S.; Qader, S.A.U. Purification, characterization and end product analysis of dextran degrading endodextranase from bacillus licheniformis kibge-ib25. Int. J. Biol. Macromol. 2015, 78, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Cai, R.; Yan, W.; Lyu, M.; Fang, Y.; Wang, S. Purification and characterization of a biofilm-degradable dextranase from a marine bacterium. Mar. Drugs 2018, 16, 51. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Nam, S.H.; Park, H.J.; Kim, Y.M.; Kim, N.; Kim, G.; Seo, E.S.; Kang, S.S.; Kim, D. Biochemical characterization of dextranase from Arthrobacter oxydans and its cloning and expression in Escherichia coli. Food Sci. Biotechnol. 2010, 19, 757–762. [Google Scholar] [CrossRef]
- Tsutsumi, K.; Gozu, Y.; Nishikawa, A.; Tonozuka, T. Structural insights into polysaccharide recognition by flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31. FEBS J. 2019, 287, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, E.R. Dextranase in sugar industry: A review. Sugar Tech 2009, 11, 124–134. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, N.; Tian, Y. Characterization and application of dextranase produced by chaetomium globosum mutant through combined application of atmospheric and room temperature plasma and ethyl methyl sulfone. Process Biochem. 2019, 85, 116–124. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Seo, M.-Y.; Kang, H.-K.; Atsuo, K.; Kim, D. Construction of a fusion enzyme of dextransucrase and dextranase: Application for one-step synthesis of isomaltooligosaccharides. Enzym. Microb. Technol. 2009, 44, 159–164. [Google Scholar] [CrossRef]
- Chen, Y.; Qian, J.; Zhao, C.; Yang, L.; Ding, J.; Guo, H. Preparation and evaluation of porous starch/chitosan composite cross-linking hemostatic. Eur. Polym. J. 2019, 118, 17–26. [Google Scholar] [CrossRef]
- Dong, D.; Wang, X.; Deng, T.; Ning, Z.; Tian, X.; Zu, H.; Ding, Y.; Wang, C.; Wang, S.; Lyu, M. A novel dextranase gene from the marine bacterium Bacillus aquimaris s5 and its expression and characteristics. FEMS Microbiol. Lett. 2021, 368. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Liu, N.; Wei, Z.; Wang, Z.; Wang, Y.; Wang, S. Purification and characterization of cold-adapted and salt-tolerant dextranase from Cellulosimicrobium sp. thn1 and its potential application for treatment of dental plaque. Front. Microbiol. 2022, 13, 1012957. [Google Scholar] [CrossRef] [PubMed]
- Ebaya, M.M.A.; El-Mowafy, M.; Adel El-Sokkary, M.M.; Hassan, R. Purification, characterization, and biocatalytic and antibiofilm activity of a novel dextranase from Talaromyces sp. Int. J. Microbiol. 2020, 2020, 9198048. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, H.; Lu, M.; Fang, Y.; Jiao, Y.; Li, W.; Zhao, G.; Wang, S. Dextranase from arthrobacter oxydans kq11-1 inhibits biofilm formation by polysaccharide hydrolysis. Biofouling 2016, 32, 1223–1233. [Google Scholar] [CrossRef]
- Ning, Z.; Dong, D.; Tian, X.; Zu, H.; Tian, X.; Lyu, M.; Wang, S. Alkalic dextranase produced by marine bacterium Cellulosimicrobium sp. px02 and its application. J. Basic Microbiol. 2021, 61, 1002–1015. [Google Scholar] [CrossRef]
- Sajilata, M.G.; Singhal, R.S.; Kulkarni, P.R. Resistant starch a review. Compr. Rev. Food Sci. Food Saf. 2006, 5, 1–17. [Google Scholar] [CrossRef]
- You, H.; Liang, C.; Zhang, O.; Xu, H.; Xu, L.; Chen, Y.; Xiang, X. Variation of resistant starch content in different processing types and their starch granules properties in rice. Carbohydr. Polym. 2022, 276, 118742. [Google Scholar] [CrossRef]
- Benavent-Gil, Y.; Rosell, C.M. Morphological and physicochemical characterization of porous starches obtained from different botanical sources and amylolytic enzymes. Int. J. Biol. Macromol. 2017, 103, 587–595. [Google Scholar] [CrossRef]
- Dura, A.; Rosell, C.M. Physico-chemical properties of corn starch modified with cyclodextrin glycosyltransferase. Int. J. Biol. Macromol. 2016, 87, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Purwadi, R.; Lestari, D.; Lohoo, C.A.; Tirtaadji, J.L. The effect of size and solid content in hydrolysis of sweet potato starch using endogenous beta-amylase enzyme. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1143, 012043. [Google Scholar] [CrossRef]
- Offiah, V.; Kontogiorgos, V.; Falade, K.O. Extrusion processing of raw food materials and by-products: A review. Crit. Rev. Food Sci. Nutr. 2018, 59, 2979–2998. [Google Scholar] [CrossRef]
- Soykeabkaew, N.; Thanomsilp, C.; Suwantong, O. A review: Starch-based composite foams. Compos. Part A Appl. Sci. Manuf. 2015, 78, 246–263. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Goodfellow, M. Biosystematics of alkaliphilic streptomycetes isolated from seven locations across a beach and dune sand system. Antonie Van Leeuwen-Hoek 2008, 94, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Wang, L.; Luo, Y. Isolation, identification, and the growth promoting effects of two antagonistic actinomycete strains from the rhizosphere of mikania micrantha kunth. Microbiol. Res. 2018, 208, 1–11. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Jiao, Y.-L.; Wang, S.-J.; Lv, M.-S.; Jiao, B.H.; Li, W.J.; Fang, Y.W.; Liu, S. Characterization of a marine-derived dextranase and its application to the prevention of dental caries. J. Ind. Microbiol. Biotechnol. 2013, 41, 17–26. [Google Scholar] [CrossRef]
- Lei, M.; Jiang, F.-C.; Cai, J.; Hu, S.; Zhou, R.; Liu, G.; Wang, Y.H.; Wang, H.B.; He, J.R.; Xiong, X.G. Facile microencapsulation of olive oil in porous starch granules: Fabrication, characterization, and oxidative stability. Int. J. Biol. Macromol. 2018, 111, 755–761. [Google Scholar] [CrossRef]

| Reagents | Relative Activity (%) | ||
|---|---|---|---|
| 1 mM | 5 mM | 10 mM | |
| Control | 100.00 ± 0.08 | 100.00 ± 0.08 | 100.00 ± 0.08 |
| Ca2+ | 92.40 ± 0.56 | 102.42 ± 0.24 | 108.00 ± 0.42 |
| Ba2+ | 90.21 ± 0.08 | 85.42 ± 0.21 | 81.52 ± 0.06 |
| Mg2+ | 90.00 ± 0.12 | 86.21 ± 0.54 | 80.24 ± 0.24 |
| Co2+ | 75.42 ± 0.48 | 50.21 ± 0.06 | 20.84 ± 0.56 |
| Sr2+ | 95.46 ± 1.02 | 105.26 ± 0.82 | 124.25 ± 0.54 |
| Zn2+ | 85.24 ± 0.82 | 72.21 ± 0.24 | 50.56 ± 0.42 |
| K+ | 98.40 ± 2.20 | 104.10 ± 1.52 | 112.62 ± 0.20 |
| Fe3+ | 72.32 ± 0.47 | 0.00 | 0.00 |
| Substrate | Main Linkages | Relative Activity (%) |
|---|---|---|
| Dextran T20 | α-1,6 | 100.00 ± 0.25 |
| Dextran T40 | α-1,6 | 95.00 ± 0.39 |
| Dextran T70 | α-1,6 | 90.00 ± 0.72 |
| Dextran T500 | α-1,6 | 85.00 ± 0.46 |
| Dextran T2000 | α-1,6 | 80.00 ± 0.52 |
| Soluble starch | α-1,4; α-1,6 | 32.00 ± 0.45 |
| Chitin | β-1,4 | 0 |
| Time of Hydrolysis (h) | Hydrolysates (%) | |||
|---|---|---|---|---|
| Isomaltoheptaose | Isomaltopentaose | Maltose | Glucose | |
| 0.5 | 98.02 | 1.81 | 0.06 | 0.10 |
| 2 | 97.18 | 2.18 | 0.52 | 0.12 |
| 6 | 72.71 | 22.43 | 0.53 | 4.33 |
| Concentration of Dextranase (U/mL) | Biofilm Formation Inhibition Rate (%) | Formed Biofilm Reduction Rate (%) |
|---|---|---|
| 0 | 0.00 | 0.00 |
| 2 | 27.32 ± 0.56 | 50.12 ± 2.34 |
| 4 | 52.15 ± 2.24 | 65.14 ± 1.21 |
| 6 | 75.82 ± 1.25 | 73.28 ± 0.35 |
| 8 | 83.26 ± 2.12 | 79.24 ± 2.41 |
| 10 | 94.23 ± 0.22 | 92.54 ± 1.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Wu, Y.; Li, Q.; Wu, X.; Kang, X.; Zhang, L.; Lyu, M.; Wang, S. The Screening and Identification of a Dextranase-Secreting Marine Actinmycete Saccharomonospora sp. K1 and Study of Its Enzymatic Characteristics. Mar. Drugs 2024, 22, 69. https://doi.org/10.3390/md22020069
Wang B, Wu Y, Li Q, Wu X, Kang X, Zhang L, Lyu M, Wang S. The Screening and Identification of a Dextranase-Secreting Marine Actinmycete Saccharomonospora sp. K1 and Study of Its Enzymatic Characteristics. Marine Drugs. 2024; 22(2):69. https://doi.org/10.3390/md22020069
Chicago/Turabian StyleWang, Boyan, Yizhuo Wu, Qiang Li, Xudong Wu, Xinxin Kang, Lei Zhang, Mingsheng Lyu, and Shujun Wang. 2024. "The Screening and Identification of a Dextranase-Secreting Marine Actinmycete Saccharomonospora sp. K1 and Study of Its Enzymatic Characteristics" Marine Drugs 22, no. 2: 69. https://doi.org/10.3390/md22020069





