High Structural Diversity of Aeruginosins in Bloom-Forming Cyanobacteria of the Genus Planktothrix as a Consequence of Multiple Recombination Events
Abstract
:1. Introduction
2. Results and Discussion
2.1. Aeruginosin Biosynthesis Genes Show Multiple Recombinations and a Variable Evolutionary Origin
2.2. Aer Gene Organization and Composition in Relation to Planktothrix Evolution
2.3. Aeruginosin Structural Variation and Chemical Diversification in Relation to Phylogenetic Lineages
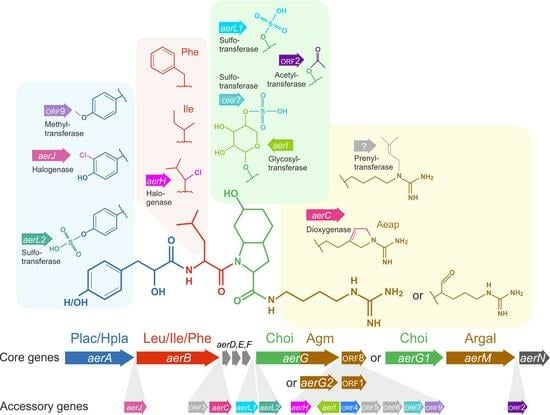
2.4. Relationships between Aeruginosin Structural Modification and Core/Accessory Aer Genes
2.5. High Resolution Mass Spectrometry (HRMS)
2.6. Toxicity of Aeruginosins Resulting from the Large-Range Recombination Events
3. Conclusions
4. Materials and Methods
4.1. Organisms
4.2. DNA Extraction, PCR and Sequencing
4.3. Sequence Comparison and Phylogenetic Analysis
4.4. Multivariate Statistical Analysis
4.5. Aeruginosin Peptide Identification and Fragmentation Using HPLC-MSn
4.6. Aeruginosin Peptide Purification and Structure Identification Using HRMS
4.7. Toxicity Tests
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, I.S.; Pietrasiak, N.; Gobler, C.J.; Johansen, J.R.; Burkholder, J.M.; D’Antonio, S.; Zimba, P.V. Diversity of bioactive compound content across 71 genera of marine, freshwater, and terrestrial cyanobacteria. Harmful Algae 2021, 109, 102116. [Google Scholar] [CrossRef] [PubMed]
- Ersmark, K.; Del Valle, J.R.; Hanessian, S. Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew. Chem. Int. Ed. Engl. 2008, 47, 1202–1223. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, M.; Huang, Z.; Fang, J.; Wang, Z.; Zhou, C.; Qiu, X. Diversity, biosynthesis and bioactivity of aeruginosins, a family of cyanobacteria-derived nonribosomal linear tetrapeptides. Mar. Drugs 2023, 21, 217. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, M.; Maruthanayagam, V.; Sundararaman, M. SAR analysis and bioactive potentials of freshwater and terrestrial cyanobacterial compounds: A review. J. Appl. Toxicol. 2013, 33, 313–349. [Google Scholar] [CrossRef]
- Wang, G.; Goyal, N. Aeruginosin analogs and other compounds with rigid bicyclic structure as potential antithrombotic agents. Cardiovasc. Hematol. Agents Med. Chem. 2009, 7, 147–165. [Google Scholar] [CrossRef]
- Kurmayer, R.; Deng, L.; Entfellner, E. Role of toxic and bioactive secondary metabolites in colonization and bloom formation by filamentous cyanobacteria Planktothrix. Harmful Algae 2016, 54, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, M.; Kobos, J.; Toruńska-Sitarz, A.; Mazur-Marzec, H. Non-ribosomal peptides produced by Planktothrix agardhii from Siemianówka Dam Reservoir SDR (northeast Poland). Arch. Microbiol. 2014, 196, 697–707. [Google Scholar] [CrossRef]
- Toporowska, M.; Mazur-Marzec, H.; Pawlik-Skowrońska, B. The effects of cyanobacterial bloom extracts on the biomass, Chl-a, MC and other oligopeptides contents in a natural Planktothrix agardhii Population. Int. J. Environ. Res. Public Health 2020, 17, 2881. [Google Scholar] [CrossRef]
- Welker, M.; von Döhren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef]
- Stenico, E.; Rigonato, J.; Lorenzi, A.S.; Azevedo, M.T.; Sant’Anna, C.L.; Fiore, M.F. Bioactive cyanopeptides produced by Sphaerocavum brasiliense strains (Cyanobacteria). J. Braz. Chem. Soc. 2015, 26, 2088–2096. [Google Scholar] [CrossRef]
- Qiu, X.; Zhu, W.; Wang, W.; Jin, H.; Zhu, P.; Zhuang, R.; Yan, X. Structural and functional insights into the role of a cupin superfamily isomerase in the biosynthesis of Choi moiety of aeruginosin. J. Struct. Biol. 2019, 205, 44–52. [Google Scholar] [CrossRef]
- Qiu, X.; Wei, Y.; Zhu, W.; Fu, J.; Duan, X.; Jin, H.; Zhu, P.; Zhou, C.; Yan, X. Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin. J. Struct. Biol. 2020, 209, 107415. [Google Scholar] [CrossRef]
- Ishida, K.; Christiansen, G.; Yoshida, W.Y.; Kurmayer, R.; Welker, M.; Valls, N.; Bonjoch, J.; Hertweck, C.; Börner, T.; Hemscheidt, T.; et al. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 2007, 14, 565–576. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Wahlsten, M.; Jokela, J.; Nees, M.; Stenman, U.H.; Alvarenga, D.O.; Strandin, T.; Sivonen, K.; Poso, A.; Permi, P.; et al. Potent inhibitor of human trypsins from the aeruginosin family of natural products. ACS Chem. Biol. 2021, 16, 2537–2546. [Google Scholar] [CrossRef]
- Calteau, A.; Fewer, D.P.; Latifi, A.; Coursin, T.; Laurent, T.; Jokela, J.; Kerfeld, C.A.; Sivonen, K.; Piel, J.; Gugger, M. Phylum-wide comparative genomics unravel the diversity of secondary metabolism in cyanobacteria. BMC Genom. 2014, 15, 977. [Google Scholar] [CrossRef]
- Schorn, M.A.; Jordan, P.A.; Podell, S.; Blanton, J.M.; Agarwal, V.; Biggs, J.S.; Allen, E.E.; Moore, B.S. Comparative genomics of cyanobacterial symbionts reveals distinct, specialized metabolism in tropical Dysideidae sponges. mBio 2019, 10, e00821-19. [Google Scholar] [CrossRef] [PubMed]
- Heinilä, L.M.P.; Jokela, J.; Ahmed, M.N.; Wahlsten, M.; Kumar, S.; Hrouzek, P.; Permi, P.; Koistinen, H.; Fewer, D.P.; Sivonen, K. Discovery of varlaxins, new aeruginosin-type inhibitors of human trypsins. Org. Biomol. Chem. 2022, 20, 2681–2692. [Google Scholar] [CrossRef]
- Fewer, D.P.; Jokela, J.; Paukku, E.; Österholm, J.; Wahlsten, M.; Permi, P.; Aitio, O.; Rouhiainen, L.; Gomez-Saez, G.V.; Sivonen, K. New structural variants of aeruginosin produced by the toxic bloom forming cyanobacterium Nodularia spumigena. PLoS ONE 2013, 8, e73618. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Budnjo, A.; Jokela, J.; Haug, B.E.; Fewer, D.P.; Wahlsten, M.; Rouhiainen, L.; Permi, P.; Fossen, T.; Sivonen, K. Pseudoaeruginosins, nonribosomal peptides in Nodularia spumigena. ACS Chem. Biol. 2015, 10, 725–733. [Google Scholar] [CrossRef]
- Fujii, K.; Sivonen, K.; Adachi, K.; Noguchi, K.; Shimizu, Y.; Sano, H.; Hirayama, K.; Suzuki, M.; Harada, K.-I. Comparative study of toxic and non-toxic cyanobacterial products: A novel glycoside, suomilide, from non-toxic Nodularia spumigena HKVV. Tetrahedron Lett. 1997, 38, 5529–5532. [Google Scholar] [CrossRef]
- Ishida, K.; Welker, M.; Christiansen, G.; Cadel-Six, S.; Bouchier, C.; Dittmann, E.; Hertweck, C.; Tandeau de Marsac, N. Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Appl. Environ. Microbiol. 2009, 75, 2017–2026. [Google Scholar] [CrossRef] [PubMed]
- Shimura, Y.; Fujisawa, T.; Hirose, Y.; Misawa, N.; Kanesaki, Y.; Nakamura, Y.; Kawachi, M. Complete sequence and structure of the genome of the harmful algal bloom-forming cyanobacterium Planktothrix agardhii NIES-204(T) and detailed analysis of secondary metabolite gene clusters. Harmful Algae 2021, 101, 101942. [Google Scholar] [CrossRef] [PubMed]
- Entfellner, E.; Frei, M.; Christiansen, G.; Deng, L.; Blom, J.; Kurmayer, R. Evolution of anabaenopeptin peptide structural variability in the cyanobacterium Planktothrix. Front. Microbiol. 2017, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Kurmayer, R.; Blom, J.F.; Deng, L.; Pernthaler, J. Integrating phylogeny, geographic niche partitioning and secondary metabolite synthesis in bloom-forming Planktothrix. ISME J. 2015, 9, 909–921. [Google Scholar] [CrossRef] [PubMed]
- McKindles, K.M.; McKay, R.M.; Bullerjahn, G.S. Genomic comparison of Planktothrix agardhii isolates from a Lake Erie embayment. PLoS ONE 2022, 17, e0273454. [Google Scholar] [CrossRef] [PubMed]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2—A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed]
- Pancrace, C.; Barny, M.A.; Ueoka, R.; Calteau, A.; Scalvenzi, T.; Pédron, J.; Barbe, V.; Piel, J.; Humbert, J.F.; Gugger, M. Insights into the Planktothrix genus: Genomic and metabolic comparison of benthic and planktic strains. Sci. Rep. 2017, 7, 41181. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Ishida, K.; Okino, T.; Okita, Y.; Matsuda, H.; Yamaguchi, K. Aeruginosins 98-A and B, trypsin inhibitors from the blue-green alga Microcystis aeruginosa (NIES-98). Tetrahedron Lett. 1995, 36, 2785–2788. [Google Scholar] [CrossRef]
- Cadel-Six, S.; Dauga, C.; Castets, A.M.; Rippka, R.; Bouchier, C.; Tandeau de Marsac, N.; Welker, M. Halogenase genes in nonribosomal peptide synthetase gene clusters of Microcystis (cyanobacteria): Sporadic distribution and evolution. Mol. Biol. Evol. 2008, 25, 2031–2041. [Google Scholar] [CrossRef]
- Kohler, E.; Grundler, V.; Häussinger, D.; Kurmayer, R.; Gademann, K.; Pernthaler, J.; Blom, J.F. The toxicity and enzyme activity of a chlorine and sulfate containing aeruginosin isolated from a non-microcystin-producing Planktothrix strain. Harmful Algae 2014, 39, 154–160. [Google Scholar] [CrossRef]
- Ishida, K.; Okita, Y.; Matsuda, H.; Okino, T.; Murakami, M. Aeruginosins, protease inhibitors from the cyanobacterium Microcystis aeruginosa. Tetrahedron 1999, 55, 10971–10988. [Google Scholar] [CrossRef]
- Elkobi-Peer, S.; Carmeli, S. New prenylated aeruginosin, microphycin, anabaenopeptin and micropeptin analogues from a Microcystis bloom material collected in Kibbutz Kfar Blum, Israel. Mar. Drugs 2015, 13, 2347–2375. [Google Scholar] [CrossRef]
- Scherer, M.; Bezold, D.; Gademann, K. Investigating the toxicity of the aeruginosin chlorosulfopeptides by chemical synthesis. Angew. Chem. Int. Ed. Engl. 2016, 55, 9427–9431. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988, 167, 3–27. [Google Scholar] [CrossRef] [PubMed]
- Suda, S.; Watanabe, M.M.; Otsuka, S.; Mahakahant, A.; Yongmanitchai, W.; Nopartnaraporn, N.; Liu, Y.; Day, J.G. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 2002, 52 Pt 5 Pt 5, 1577–1595. [Google Scholar] [CrossRef]
- Gaget, V.; Welker, M.; Rippka, R.; de Marsac, N.T. A polyphasic approach leading to the revision of the genus Planktothrix (Cyanobacteria) and its type species, P. agardhii, and proposal for integrating the emended valid botanical taxa, as well as three new species, Planktothrix paucivesiculata sp. nov. ICNP, Planktothrix tepida sp. nov. ICNP, and Planktothrix serta sp. nov. ICNP, as genus and species names with nomenclatural standing under the ICNP. Syst. Appl. Microbiol. 2015, 38, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kosol, S.; Schmidt, J.; Kurmayer, R. Variation in peptide net production and growth among strains of the toxic cyanobacterium Planktothrix spp. Eur. J. Phycol. 2009, 44, 49–62. [Google Scholar] [CrossRef]
- Törökné, A. Thamnocephalus Test. In Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; Meriluoto, J., Spoof, L., Codd, G., Eds.; John Wiley & Sons, Ltd.: Chichester, West Sussex, PO19 8SQ, UK, 2017; pp. 462–468. [Google Scholar]
- Kodani, S.; Ishida, K.; Murakami, M. Aeruginosin 103-A, a thrombin inhibitor from the cyanobacterium Microcystis viridis. J. Nat. Prod. 1998, 61, 1046–1048. [Google Scholar] [CrossRef]
- Harada, K.-I.; Fujii, K.; Shimada, T.; Suzuki, M.; Sano, H.; Adachi, K.; Carmichael, W.W. Two cyclic peptides, anabaenopeptins, a third group of bioactive compounds from the cyanobacterium Anabaena flos-aquae NRC 525-17. Tetrahedron Lett. 1995, 36, 1511–1514. [Google Scholar] [CrossRef]
- Blom, J.F.; Robinson, J.A.; Jüttner, F. High grazer toxicity of [D-Asp(3),(E)-Dhb(7)]microcystin-RR of Planktothrix rubescens as compared to different microcystins. Toxicon 2001, 39, 1923–1932. [Google Scholar] [CrossRef] [PubMed]




| Gene | Functional Description | Literature |
|---|---|---|
| aerA | NRPS/PKS hydrid (incorporation of Plac/Hpla at pos. 1) | [13,21] |
| aerB | NRPS (incorporation of Leu/Ile/Phe at pos. 2) | [13,21] |
| aerC | oxygenase (oxidation of Agm to Aeap) | [14]; this study 1 |
| aerD, E, F | Choi synthesis (isomerase, reductase) | [11,12,13] |
| aerG, G1 + G2, G1 + M | NRPS (Choi at pos. 3 and Agm derivate at pos. 4) | [13,21] |
| aerH | halogenase (chlorination at Leu at pos. 2) | this study |
| aerI | glycosyltransferase (Xyl at Choi) | [13,21] |
| aerJ | halogenase (chlorination at Hpla at pos. 1) | [21,28,29]; this study |
| aerK | isomerase | [14] |
| aerL1 | sulfotransferase (sulfation at Choi at pos. 3) | [21]; this study |
| aerL2 | sulfotransferase (sulfation at Hpla at pos. 1) | [21]; this study |
| aerN | ABC transporter | [13,21] |
| ORF1 | oxidoreductase (reduction of Agm; cleavage from synthesis operon) | [14]; this study |
| ORF2 | hypothetical protein (O-acetylation at Choi) | this study |
| ORF3 | hypothetical protein | this study |
| ORF4 | sulfotransferase (sulfation at unknown pos.) | [14]; this study |
| ORF5 | hypothetical protein | |
| ORF6 | hypothetical protein | |
| ORF7 | sulfotransferase (sulfation at Xyl) | [14]; this study |
| ORF8 | aldo-/keto reductase (reduction of Agm; cleavage from synthesis operon) | [14]; this study |
| ORF9 | hypothetical protein (O-methylation at Hpla at pos. 1) | this study |
| Strain |
Retention Time [min] | [M+H]+ |
Choi Fragment [M+H]+ | Chlorine Detected | Sum Formula |
|---|---|---|---|---|---|
| NIVA-CYA116 | 4.1 | 717.25 | 140.1 | Yes | nd |
| 5.9 | 689.25 | 140.1 | Yes | nd | |
| 12.5 | 759.26 | 122.1 | Yes | nd | |
| 16.5 | 731.28 | 122.1 | Yes | nd | |
| No1020 | 6.6 | 717.25 | 140.1 | Yes | nd |
| No66 | 6.3 | 717.20 | 140.1 | Yes | C30H46N6O10SCl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Entfellner, E.; Baumann, K.B.L.; Edwards, C.; Kurmayer, R. High Structural Diversity of Aeruginosins in Bloom-Forming Cyanobacteria of the Genus Planktothrix as a Consequence of Multiple Recombination Events. Mar. Drugs 2023, 21, 638. https://doi.org/10.3390/md21120638
Entfellner E, Baumann KBL, Edwards C, Kurmayer R. High Structural Diversity of Aeruginosins in Bloom-Forming Cyanobacteria of the Genus Planktothrix as a Consequence of Multiple Recombination Events. Marine Drugs. 2023; 21(12):638. https://doi.org/10.3390/md21120638
Chicago/Turabian StyleEntfellner, Elisabeth, Kathrin B. L. Baumann, Christine Edwards, and Rainer Kurmayer. 2023. "High Structural Diversity of Aeruginosins in Bloom-Forming Cyanobacteria of the Genus Planktothrix as a Consequence of Multiple Recombination Events" Marine Drugs 21, no. 12: 638. https://doi.org/10.3390/md21120638