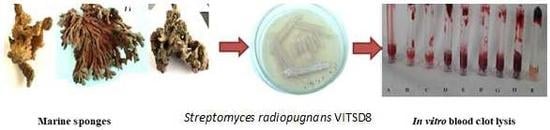
Novel Fibrinolytic Protease Producing Streptomyces radiopugnans VITSD8 from Marine Sponges
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Collection and Isolation of Actinomycetes
2.2.1. Collection of Sponges
2.2.2. Spicule Preparation
2.2.3. Isolation and Screening for Streptomyces sp. from Marine Sponges
2.3. Bacterial Strain and Culture Conditions
2.4. Determination of Protease Activity by Caseinolytic Method
2.5. Quantification of Protein and Enzyme Activity
2.6. Characterization of Selected Streptomyces sp.
2.6.1. Molecular and Phylogenetic Analysis
2.6.2. Scanning Electron Microscope (SEM) Analysis
2.7. Optimization
2.8. Purification
2.9. Blood Clot Lysis Assay
2.10. Fibrin Plate Method
2.11. Partial Clot Lysis Assay
2.12. HPLC Analysis
2.13. FTIR Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boersma, E.; Mercado, N.; Poldermans, D.; Gardien, M.; Vos, J.; Simoons, M.L. Acute myocardial infarction. Lancet 2003, 361, 847–858. [Google Scholar] [CrossRef]
- Blix, S. The effectiveness of activators in clot lysis, with special reference to fibrinolytic therapy: A new method for determination of preformed clot lysis. Acta Med. Scand. 1962, 172, 1–24. [Google Scholar] [CrossRef]
- Sumi, H.; Hamada, H.; Tsushima, H.; Mihara, H.; Muraki, H. A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia 1987, 43, 1110–1111. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, V.G.; Kirklin, J.K.; Holman, W.L.; Steenwyk, B.L. Clot lifespan model analysis of the effects of warfarin on thrombus growth and fibrinolysis: Role of contact protein and tissue factor initiation. ASAIO J. 2009, 55, 33–40. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, S.; Mitova, M.; Tommonaro, G. Marine bacteria associated with sponge as source of cyclic peptides. Biomol. Eng. 2003, 20, 311–316. [Google Scholar] [CrossRef]
- Maya, K. Studies on L-asparaginase activity in Bacillus of retting ground. Fish Technol. 1992, 29, 62–66. [Google Scholar] [CrossRef]
- Ratnakar, R.C.; Siddharath, V.D. Production, purification and biochemical characterization of a fibrinolytic enzyme from thermophilic Streptomyces sp. MCMB-379. Appl. Biochem. Biotechnol. 2011, 165, 1406–1413. [Google Scholar] [CrossRef]
- Dilip, C.V.; Mulaje, S.S.; Mohalkar, R.Y. A review on actinomycetes and their biotechnological application. Int. J. Biopharm. Sci. Res. 2013, 4, 1730–1742. [Google Scholar] [CrossRef]
- Mohanasrinivasan, V.; Yogesh, S.; Govindaraj, A.; Naine, J.S.; Devi, S.C. In vitro thrombolytic potential of actinoprotease from marine Streptomyces violaceus VITYGM. Cardiovasc. Hematol. Agents Med. Chem. 2016, 14, 120–124. [Google Scholar] [CrossRef]
- Naveena, B.; Gopinath, K.P.; Sakthiselvan, P.; Partha, N. Enhanced production of thrombinase by Streptomyces venezuelae: Kinetic studies on growth and enzyme production of mutant strain. Bioresour. Technol. 2012, 1, 417–424. [Google Scholar] [CrossRef]
- Best, M.; Kenchington, E.; MacIsaac, K.; Wareham, V.; Fuller, S.D.; Thompson, A.B. Sponge identification guide NAFO area. NAFO Sci. Coun. Stud. 2010, 43, 1–49. [Google Scholar] [CrossRef]
- Hooper, J.N. Sponguide: Guide to sponge collection and identification. Qld. Mus. 2000. [Google Scholar] [CrossRef]
- Van Soest, R.W.; Hooper, J.N. Systema Porifera: A Guide to the Classification of Sponges; Kluwer Academic/Plenum: Boston, MA, USA, 2002. [Google Scholar] [CrossRef]
- Montalvo, N.F.; Mohamed, N.M.; Enticknap, J.J.; Hill, R.T. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef]
- Egorov, N.S.; Prianishnikova, N.I.; Aslanian, R.R. Streptomyces spheroides M8-2 strain—A producer of extracellular proteolytic enzymes possessing fibrinolytic and thrombolytic action. Nauchnye Dokl. Vyss. Shkoly Biol. Nauki 1985, 1, 77–81. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Oda, K.; Murao, S. Purification and some enzymatic properties of acid protease A and B of Scytalidium lignicolum ATCC 24568. Agric. Biol. Chem. 1974, 38, 2435–2444. [Google Scholar] [CrossRef]
- Locci, R. Streptomyces and related genera. Bergey’s Man. Syst. Bacteriol. 1989, 4, 2451–2508. [Google Scholar] [CrossRef]
- Magarvey, N.A.; Keller, J.M.; Bernan, V.; Dworkin, M.; Sherman, D.H. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl. Environ. Microbiol. 2004, 70, 7520–7529. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.T.; Davies, F.L. Use of a scanning electron microscope for the examination of actinomycetes. Microbiology 1967, 48, 171–177. [Google Scholar] [CrossRef]
- Xiuyunju, X.C.; Yong, S.; Zhe, W.; Chengliang, C.; Jinjuan, L.; Jihong, J. Purification and characterization of a fibrinolytic enzyme from Streptomyces sp. XZNUM 00004. World J. Microbial. Biotechnol. 2012, 28, 2479–2486. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680. [Google Scholar] [CrossRef]
- Abdel-Naby, M.A.; El-Diwany, A.I.; Shaker, H.M.; Ismail, A.M. Production and properties of fibrinolytic enzyme from Streptomyces sp. NRC 411. World J. Microbiol. Biotechnol. 1992, 8, 267–269. [Google Scholar] [CrossRef]
- Astrup, T.; Müllertz, S. The fibrin plate method for estimating fibrinolytic activity. Arch. Biochem. Biophys. 1952, 40, 346–351. [Google Scholar] [CrossRef]
- Haris, P.I.; Severcan, F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B-Enzym. 1999, 7, 207–221. [Google Scholar] [CrossRef]
- Carter, H.J. Descriptions and figures of deep-sea sponges and their spicules from the Atlantic Ocean, dredged up on board H.M.S. ‘Porcupine’, chiefly in 1869; with figures and descriptions of some remarkable Spicules from the Agulhas Shoal and Colon, Panama. Ann. Mag. Nat. Hist. 1874, 14, 207–221, 245–257. [Google Scholar] [CrossRef]
- Cheng, G.; He, L.; Sun, Z.; Cui, Z.; Du, Y.; Kong, Y. Purification and biochemical characterization of a novel fibrinolytic enzyme from Streptomyces sp. P3. J. Microbiol. Biotechnol. 2015, 25, 1449–1459. [Google Scholar] [CrossRef]
- Chitte, R.R.; Dey, S. Potent fibrinolytic enzyme from a thermophilic Streptomyces megasporus strain SD5. Lett. Appl. Microbiol. 2000, 31, 405–410. [Google Scholar] [CrossRef]
- Velumani, S. Isolation, screening, characterization and production of fibrinolytic enzyme from marine microorganism. Int. J. Adv. Res. 2016, 2, 2395–4396. [Google Scholar] [CrossRef]
- Hanan, M.M.; Abd, E.A.; Babiker, M.B.; Aisha, E.A.B. Isolation and characterization of thermophilic Streptomyces sp. with potential production of actinokinase. Int. J. Adv. Res. 2016, 4, 869–878. [Google Scholar] [CrossRef]
- Gesheva, V. Production of fibrinolytic enzyme by Streptomyces rimosus at conditions of nitrogen limitation. J. Microbiol. Biochem. Tech. 2009, 1, 57–58. [Google Scholar] [CrossRef]
- Verma, P.; Chatterjee, S.; Merlyn Keziah, S.; Devi, S.C. Fibrinolytic protease from marine Streptomyces rubiginosus VITPSS1. Cardiovasc. Hematol. Agents Med. Chem. 2018, 16, 44–45. [Google Scholar] [CrossRef]











| Microscopy | Corn Steep Liquor | + | |
|---|---|---|---|
| Grams Stain | + | Sodium acetate | + |
| Colony Pigmentation | - | Sodium citrate | - |
| Melanin Pigmentation | - | Urea | - |
| Motility | - | Effect of Temperature | |
| Soluble Pigment | - | 15 °C | - |
| Carbon Source | - | 28 °C | + |
| D-glucose | + | 37 °C | + |
| D-galactose | - | 45 °C | - |
| Mannose | + | Effect of pH | |
| Maltose | + | 5 | - |
| Lactose | + | 6 | - |
| Trehalose | + | 7 | + |
| Melibose | - | 8 | - |
| Amino Acid Utilization | 9 | - | |
| Cysteine | + | NaCl Concentration | |
| Arginine | + | 1% | + |
| Therionine | + | 2% | + |
| Alanine | + | 3% | - |
| Aspartic acid | + | 5% | - |
| Glycine | + | 10% | - |
| Histidine | - | Hydrolysis | |
| Lysine | + | Starch Hydrolysis | + |
| Phenyl alanine | + | Nitrate Reduction | - |
| Tryptophan | + | Gelatin Liquefaction | + |
| Methionine | + | Hemolysis on Blood Agar | + |
| Isoleucine | - | Esculin Hydrolysis | - |
| Valine | - | H2S | - |
| Ornithine | - | Antibiotic Resistance | |
| Asparagine | - | Streptomycin | S |
| Nitrogen Source | Tetracyclin | S | |
| Peptone | + | Bacitracin | R |
| Yeast Extract | + | Kanamycin | S |
| Casein | + | Ampicillin | R |
| Ammonium Sulphate | + | Gentamicin | R |
| Ammonium Nitrate | + | Rifampacin | R |
| Ammonium Citrate | + | Vancomycin | S |
| Beef Extract | + | Fluconazole | R |
| Characteristic Features | Streptomyces sp. VITSD8 | Utilization of Carbon Sources | |
|---|---|---|---|
| Spore surface | Smooth | Arabinose | ₊ |
| Spore chain morphology | Rectiflexibles | Xylose | ₋ |
| Aerial mycelium | Grey | Inositol | ₊ |
| Melanoid pigment | - | Mannitol | ₊ |
| Reverse side pigment | - | Fructose | ₊ |
| Soluble pigment | - | Rhamnose | ₊ |
| Sucrose | ₊ | ||
| Raffinose | ₊ |
| Medium | Growth | Aerial Mycelium | Substrate Mycelium |
|---|---|---|---|
| Tryptone-Yeast Extract Broth (ISP medium 1) | Good | White | Yellow |
| Yeast-malt extract (ISP medium 2) | Very good | White | Yellow |
| Oatmeal agar (ISP medium 3) | Good | White | Yellow |
| Inorganic salt starch agar (ISP medium 4) | Good | Grey | Grey |
| Glycerol-aspargine agar (ISP 5) | Good | White | Ash |
| Peptone yeast extract iron agar (ISP medium 6) | Slow | Grey | Grey |
| Tyrosine agar base (ISP medium 7) | Good | Ash | Ash |
| Nitrate agar (ISP 8) | Good | Ash | Ash |
| Carbon utilization agar (ISP medium 9) | Good | White | Grey |
| Bennett agar | Good | Grey | Grey |
| Nutrient agar | Slow | White | White |
| Czapex Dox agar | Good | White | White |
| Kenknight’s agar | Good | White | Grey |
| Actinomycete isolation agar | Very good | White | Grey |
| Starch casein agar | Very good | Grey | Grey |
| Fractions | Total Protein (mg) | Relative Activity (U) | Total Activity (U·mL) | Specific Activity (U·mg−1) | Fold Purification | % Yield |
|---|---|---|---|---|---|---|
| Crude enzyme | 71 | 247 | 12,350 | 174 | 1 | 100 |
| Ammonium sulphate precipitation | 15 | 545 | 7085 | 473 | 2.7 | 57 |
| Dialysis | 8 | 697 | 6273 | 784 | 4.51 | 51 |
| Ion-exchange chromatography | 3 | 884 | 5304 | 1768 | 10.16 | 43 |
| Size exclusion chromatography | 1.1 | 1427 | 4281 | 3891 | 22.36 | 35 |
| Time | 10 min (%) | 20 min (%) | 30 min (%) |
|---|---|---|---|
| Purified | 91 | 100 | 100 |
| Partially Purified | 57 | 78 | 86 |
| Ammonium Precipitate | 18 | 42 | 73 |
| Streptokinase | 87 | 94 | 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D, D.; S, J.N.; S, M.K.; C, S.D. Novel Fibrinolytic Protease Producing Streptomyces radiopugnans VITSD8 from Marine Sponges. Mar. Drugs 2019, 17, 164. https://doi.org/10.3390/md17030164
D D, S JN, S MK, C SD. Novel Fibrinolytic Protease Producing Streptomyces radiopugnans VITSD8 from Marine Sponges. Marine Drugs. 2019; 17(3):164. https://doi.org/10.3390/md17030164
Chicago/Turabian StyleD, Dhamodharan, Jemimah Naine S, Merlyn Keziah S, and Subathra Devi C. 2019. "Novel Fibrinolytic Protease Producing Streptomyces radiopugnans VITSD8 from Marine Sponges" Marine Drugs 17, no. 3: 164. https://doi.org/10.3390/md17030164