Quorum Sensing Inhibitors from Marine Microorganisms and Their Synthetic Derivatives
Abstract
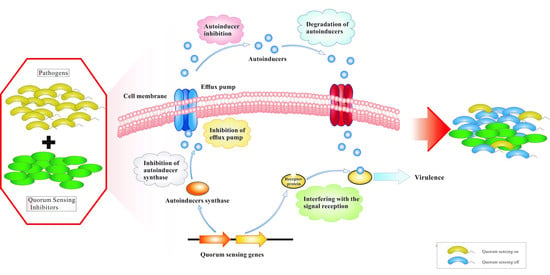
:1. Introduction
2. QSIs from Marine Bacteria and Their Derivatives with QS Inhibitory Activity
2.1. QSIs from Marine Gram-Positive Bacteria and Their Derivatives with QS Inhibitory Activity
2.2. QSIs from Marine Gram-Negative Bacteria
3. QSIs from Marine Actinomycetes and Their Derivatives with QS Inhibitory Activity
4. QSIs from Marine Fungi and Their Derivatives with QS Inhibitory Activity
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Schillaci, D.; Spanò, V.; Parrino, B.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G.; Cascioferro, S.M. Pharmaceutical approaches to target antibiotic resistance mechanisms. J. Med. Chem. 2017, 60, 8268–8297. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Parsek, M.R.; Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001, 35, 439–468. [Google Scholar] [CrossRef] [PubMed]
- Breah, L.S.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar]
- Kaufmann, G.; Park, J.; Janda, K.D. Bacterial quorum sensing: A new target for anti-infective immunotherapy. Expert Opin. Biol. Ther. 2008, 8, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.A.; Hoven, A.D.; Cook, A.M. Therapeutic frontiers: Preventing and treating infectious diseases by inhibiting bacterial quorum sensing. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 635–642. [Google Scholar] [CrossRef]
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011, 111, 28–67. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef]
- Lu, Y.; Dong, X.; Liu, S.; Bie, X. Characterization and identification of a novel marine Streptomyces sp. produced antibacterial substance. Mar. Biotechnol. 2009, 11, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Wu, Q.H.; Rowley, D.C.; Al-Kareef, A.M.; Wang, H. Anticancer agent-based marine natural products and related compounds. J. Asian Nat. Prod. Res. 2015, 17, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ashforth, E.; Ren, B.; Song, F.; Dai, H.; Liu, M.; Wang, J.; Xie, Q.; Zhang, L. Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J. Antibiot. 2010, 63, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teasdale, M.E.; Donovan, K.A.; Forschner-Dancause, S.R.; Rowley, D.C. Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar. Biotechnol. 2011, 13, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Maskey, R.P.; Asolkar, R.N.; Kapaun, E.; Wagner-Dobler, I.; Laatsch, H. Phytotoxic arylethylamides from limnic bacteria using a screening with microalgae. J. Antibiot. 2002, 55, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Teasdale, M.E.; Liu, J.; Wallace, J.; Akhlaghi, F.; Rowley, D.C. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in Gram-negative bacteria. Appl. Environ. Microbiol. 2009, 75, 567–572. [Google Scholar] [CrossRef]
- Tommonaro, G.; Abbamondi, R.; Iodice, C.; Tait, K.; De Rosa, S. Diketopiperazines produced by the halophilic Archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb. Ecol. 2012, 63, 490–495. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Dobretsov, S.; Al-Fori, M.; Gunasekera, S.P.; Sudesh, K.; Paul, V.J. Quorum-sensing inhibitory compounds from extremophilic microorganisms isolated from a hypersaline cyanobacterial mat. J. Ind. Microbiol. Biot. 2013, 40, 759–772. [Google Scholar] [CrossRef]
- Chen, X.C.; Chen, J.W.; Yan, Y.C.; Chen, S.; Xu, X.W.; Zhang, H.W.; Wang, H. Quorum sensing inhibitors from marine bacteria Oceanobacillus sp. XC22919. Nat. Prod. Res. 2018, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Saurav, K.; Bar-Shalom, R.; Haber, M.; Burgsdorf, I.; Oliviero, G.; Costantino, V.; Morgenstern, D.; Steindler, L. In search of alternative antibiotic drugs: Quorum-quenching activity in sponges and their bacterial isolates. Front. Microbiol. 2016, 7, 416. [Google Scholar] [CrossRef]
- Mansson, M.; Nielsen, A.; Kjærulff, L.; Gotfredsen, C.H.; Wietz, M.; Ingmer, H.; Gram, L.; Larsen, T.O. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine Photobacterium. Mar. Drugs 2011, 9, 2537–2552. [Google Scholar] [CrossRef]
- Richard, P.N. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2010, 48, 1429–1449. [Google Scholar]
- Ji, G.Y.; Beavis, R.; Novick, R.P. Bacterial interference caused by autoinducing peptide variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef] [PubMed]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.L.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Anita, N.; Maria, M.N.; Bojer, M.S.; Lone, G.; Larsen, T.O.; Novick, R.P.; Dorte, F.; Hanne, F.K.R.; Hanne, I. Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLoS ONE 2014, 9, e84992. [Google Scholar] [CrossRef]
- Jarraud, S.; Lyon, G.J.; Figueiredo, A.M.; Lina, G.; Gérard, L.; Vandenesch, F.; Etienne, J.; Muir, T.W.; Novick, R.P. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 2011, 182, 6517–6522. [Google Scholar] [CrossRef]
- Hansen, A.M.; Peng, P.; Baldry, M.; Perez-Gassol, I.; Christensen, S.B.; Vinther, J.M.O.; Ingmer, H.; Franzyk, H. Lactam hybrid analogues of solonamide B and autoinducing peptides as potent S. aureus AgrC antagonists. Eur. J. Med. Chem. 2018, 152, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Kjaerulff, L.; Nielsen, A.; Mansson, M.; Gram, L.; Larsen, T.O.; Ingmer, H.; Gotfredsen, C.H. Identification of four new agr quorum sensing-interfering cyclodepsipeptides from a marine Photobacterium. Mar. Drugs 2013, 11, 5051–5062. [Google Scholar] [CrossRef]
- Padmavathi, A.R.; Abinaya, B.; Pandian, S.K. Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling 2014, 30, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Dai, X.; Sun, J.; Bu, X.; Weng, C.; Li, H.; Hu, Z. A diketopiperazine factor from Rheinheimera aquimaris QSI02 exhibits anti-quorum sensing activity. Sci. Rep. 2016, 6, 39637. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.Y.; Zhang, Y.H.; Shi, X.C.; Coenye, T.; Zhang, X.H. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef]
- Saurav, K.; Costantino, V.; Venturi, V.; Steindler, L. Quorum sensing inhibitors from the sea discovered using bacterial N-acyl-homoserine lactone-based biosensors. Mar. Drugs 2017, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Liu, P.; Gong, Q.; Wang, Y.; Wang, P.; Zhu, W. α-Pyrones from the marine-derived actinomycete Nocardiopsis dassonvillei subsp. dassonvillei XG-8-1. RSC Adv. 2013, 3, 20726–20731. [Google Scholar] [CrossRef]
- Du, Y.; Sun, J.; Gong, Q.; Wang, Y.; Fu, P.; Zhu, W. New α-pyridones with quorum-sensing inhibitory activity from diversity-enhanced extracts of a Streptomyces sp. derived from marine algae. J. Agric. Food Chem. 2018, 66, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Findlay, J.A.; Tam, W.H.J.; Krepinsky, J. The chemistry of some 6-methyl-4-hydroxy-2-pyridones. Can. J. Chem. 1978, 56, 613–616. [Google Scholar] [CrossRef] [Green Version]
- Abadi, A.; Al-Deeb, O.; Al-Afify, A.; El-Kashef, H. Synthesis of 4-alkyl(aryl)-6-aryl-3-cyano- 2(1H)-pyridinones and their 2-imino isosteres as nonsteroidal cardiotonic agents. Farmaco 1999, 54, 195–201. [Google Scholar] [CrossRef]
- Cocco, M.T.; Congiu, C.; Onnis, V. Synthesis and antitumour activity of 4-hydroxy-2-pyridone derivatives. Eur. J. Med. Chem. 2000, 35, 545–552. [Google Scholar] [CrossRef]
- Öztürk, G.; Erol, D.D.; Uzbay, T.; Aytemir, M.D. Synthesis of 4(1H)-pyridinone derivatives and investigation of analgesic and antiinflammatory activities. Farmaco 2001, 56, 251–256. [Google Scholar] [CrossRef]
- Cocco, M.T.; Congiu, C.; Onnis, V. New bis(pyridyl)methane derivatives from 4-hydroxy-2- pyridones: Synthesis and antitumoral activity. Eur. J. Med. Chem. 2003, 38, 37–47. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Böger, P. Phytotoxic sites of action for molecular design of modern herbicides (Part 2): Amino acid, lipid and cell wall biosynthesis, and other targets for future herbicides. Weed Biol. Manag. 2004, 4, 59–70. [Google Scholar] [CrossRef]
- Storck, P.; Aubertin, A.; Grierson, D.S. Tosylation/mesylation of 4-hydroxy-3-nitro-2-pyridinones as an activation step in the construction of dihydropyrido[3,4-b]benzo[f][1,4]thiazepin-1-one based anti-HIV agents. Tetrahedron Lett. 2005, 46, 2919–2922. [Google Scholar] [CrossRef]
- Evidente, A.; Fiore, M.; Bruno, G.; Sparapano, L.; Motta, A. Chemical and biological characterisation of sapinopyridione, a phytotoxic 3,3,6-trisubstituted-2,4-pyridione produced by Sphaeropsis sapinea, a toxigenic pathogen of native and exotic conifers, and its derivatives. Phytochemistry 2006, 67, 1019–1028. [Google Scholar] [CrossRef]
- Macdonald, G.E.; Puri, A.; Shilling, D.G. Interactive effect of photoperiod and fluridone on growth, reproduction, and biochemistry of dioecious hydrilla (Hydrilla Verticillata). Weed Sci. 2008, 56, 189–195. [Google Scholar] [CrossRef]
- Lopez, S.N.; Ramallo, I.A.; Sierra, M.G.; Zacchino, S.A.; Furlan, R.L.E. Chemically engineered extracts as an alternative source of bioactive natural product-like compounds. Proc. Natl. Acad. Sci. USA 2007, 104, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Ramallo, I.A.; Salazar, M.O.; Mendez, L.; Furlan, R.L.E. Chemically engineered extracts: Source of bioactive compounds. Acc. Chem. Res. 2011, 44, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.S.; Ok, K.; Kim, Y.H.; Park, H.D.; Byun, Y. Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 2913–2917. [Google Scholar] [CrossRef] [PubMed]
- Naik, D.N.; Wahidullah, S.; Meena, R.M. Attenuation of Pseudomonas aeruginosa virulence by marine invertebrate-derived Streptomyces sp. Lett. Appl. Microbiol. 2013, 56, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Rajkumari, J.; Borkotoky, S.; Murali, A.; Suchiang, K.; Mohanty, S.K.; Busi, S. Cinnamic acid attenuates quorum sensing associated virulence factors and biofilm formation in Pseudomonas aeruginosa PAO1. Biotechnol. Lett. 2018, 40, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Thimmaraju, R.; Bais, H.P. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 2008, 56, 1955–1962. [Google Scholar]
- Malheiro, J.F.; Maillard, J.Y.; Borges, F.; Simões, M. Evaluation of cinnamaldehyde and cinnamic acid derivatives in microbial growth control. Int. Biodeter. Biodegr. 2018. [Google Scholar] [CrossRef]
- Gilles, B.; Shari, C.; Ulrik, H.; Serge, V.C.; Paul, C.; Louis, M.; Hans, J.N.; Tom, C. Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS ONE 2011, 6, e16084. [Google Scholar] [CrossRef]
- Joshi, J.R.; Burdman, S.; Lipsky, A.; Yariv, S.; Yedidia, I. Plant phenolic acids affect the virulence of Pectobacterium aroidearum and P. carotovorum ssp. brasiliense via quorum sensing regulation. Mol. Plant. Pathol. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Xu, J.; Yao, Z.; Jiang, Y.; Zhou, H.; Jiang, W.; Dong, K. The anti-quorum sensing activity and bioactive substance of a marine derived Streptomyces. Biotechnol. Biotechnol. Equip. 2017, 31, 1007–1015. [Google Scholar] [CrossRef]
- Hasan, S.; Ansari, M.I.; Ahmad, A.; Mishra, M. Major bioactive metabolites from marine fungi: A Review. Bioinformation 2015, 11, 176–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Jeong, J.H.; Lee, K.T.; Rho, J.R.; Choi, H.D.; Kang, J.S.; Son, B.W. Gamma-pyrone derivatives, kojic acid methyl ethers from a marine-derived fungus Alternaria [correction of Altenaria] sp. Arch. Pharm. Res. 2003, 26, 532–534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, M.; Zhu, X.; Yu, W.; Gong, Q. Equisetin as potential quorum sensing inhibitor of Pseudomonas aeruginosa. Biotechnol. Lett. 2018, 40, 1–6. [Google Scholar] [CrossRef]
- Tang, Y.D.; Zhou, M.L.; Yu, W.G.; Gong, Q.H. Isolation and identification of quorum sensing inhibitors from a strain of marine fungus Penicillium sp. QF046. Prog. Mod. Biomed. 2014, 14, 6607–6621. [Google Scholar]
- Zheng, C.J.; Sohn, M.J.; Lee, S.; Kim, W.G. Meleagrin, a new FabI inhibitor from Penicillium chryosogenum with at least one additional mode of action. PLoS ONE 2013, 8, e78922. [Google Scholar] [CrossRef]
- Dobretsov, S.; Teplitski, M.; Bayer, M.; Gunasekera, S.; Proksch, P.; Paul, V.J. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 2011, 27, 893–905. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.D.; Zhou, L.M.; Ma, Q.Y.; Huang, S.Z.; Wang, P.; Dai, H.F.; Zhao, Y.X. Metabolites with Gram-negative bacteria quorum sensing inhibitory activity from the marine animal endogenic fungus Penicillium sp. SCS-KFD08. Arch. Pharm. Res. 2016, 40, 25–31. [Google Scholar] [CrossRef]
- Martín-Rodríguez, A.J.; Reyes, F.; Martín, J.; Pérez-Yépez, J.; León-Barrios, M.; Couttolenc, A.; Espinoza, C.; Trigos, Á.; Martín, V.S.; Norte, M.; et al. Inhibition of bacterial quorum sensing by extracts from aquatic fungi: First report from marine endophytes. Mar. Drugs 2014, 12, 5503–5526. [Google Scholar]
- Van Rij, E.T.; Wesselink, M.; Chin-A.-Woeng, T.F.C.; Bloemberg, G.V.; Lugtenberg, B.J.J. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol. Plant Microb. Interact. 2004, 17, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Van Rij, E.T.; Girard, G.; Lugtenberg, B.J.J.; Bloemberg, G.V. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 2005, 151, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Widmer, K.W.; Soni, K.A.; Hume, M.E.; Beier, R.C.; Jesudhasan, P.; Pillai, S.D. Identification of poultry meat-derived fatty acids functioning as quorum sensing signal inhibitors to autoinducer-2 (AI-2). J. Food Sci. 2007, 72, M363–M368. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.A.; Jesudhasan, P.; Cepeda, M.; Widmer, K.; Jayaprakasha, G.K.; Patil, B.S.; Hume, M.E.; Pillai, S.D. Identification of ground beef–derived fatty acid inhibitors of autoinducer-2-based cell signaling. J. Food Protect. 2008, 71, 134–138. [Google Scholar] [CrossRef]
- De Carvalho, M.P.; Abraham, W.R. Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 2012, 19, 3564–3577. [Google Scholar] [CrossRef] [PubMed]
- Tung, T.T.; Jakobsen, T.H.; Dao, T.T.; Fuglsang, A.T.; Givskov, M.; Christensen, S.B.; Nielsen, J. Fusaric acid and analogues as Gram-negative bacterial quorum sensing inhibitors. Eur. J. Med. Chem. 2016, 126, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhang, X.H. Quorum quenching agents: Resources for antivirulence therapy. Mar. Drugs 2014, 12, 3245–3282. [Google Scholar] [CrossRef]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef]
- Muhlen, S.; Dersch, P. Anti-virulence strategies to target bacterial infections. Curr. Top. Microbiol. 2015, 398, 147–183. [Google Scholar]
- Wagner, S.; Sommer, R.; Hinsberger, S.; Lu, C.; Hartmann, R.W.; Empting, M.; Titz, A. Novel strategies for the treatment of Pseudomonas aeruginosa infections. J. Med. Chem. 2016, 59, 5929–5969. [Google Scholar] [CrossRef]
- Hema, M.; Princy, S.A.; Sridharan, V.; Vinoth, P.; Balamurugan, P.; Sumana, M.N. Synergistic activity of quorum sensing inhibitor, pyrizine-2-carboxylic acid and antibiotics against multi-drug resistant V. cholerae. Rsc Adv. 2016, 6, 45938–45946. [Google Scholar] [CrossRef]
- Zhou, J.W.; Luo, H.Z.; Jiang, H.; Jian, T.K.; Chen, Z.Q.; Jia, A.Q. Hordenine, a novel quorum sensing inhibitor and anti-biofilm agent against Pseudomonas aeruginosa. J. Agric. Food Chem. 2018, 66, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Soukarieh, F.; Williams, P.; Stocks, M.J.; Cámara, M. Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: Current position and future perspectives. J. Med. Chem. 2018, 61, 10385–10402. [Google Scholar] [CrossRef]



| No. | Structures of QSIs | Source | Biosensor Microorganisms | Specific Inhibitory Activity | Ref. |
|---|---|---|---|---|---|
| 1 |  | H. salinus C42 | V. harveyi BB120 | Inhibition of violacein of C. violaceum CV026 and green fluorescent protein of E. coli JB525 | [14,15] |
| 2 |  | ||||
| 3 |  | H. hispanica SK-3 | E. coli A. tumefaciens NTL4 | Inhibition of green fluorescent protein of V. anguillarum DM27 | [16] |
| 4 |  | Marinobacter sp. SK-3 | C. violaceum CV017 | Inhibition of luminescence of E. coli pSB401 | [17] |
| 5 |  | ||||
| 6 |  | ||||
| 7 |  | ||||
| 8 |  | Oceanobacillus sp.XC22919 | C. violaceum CV026 | Inhibition of violacein of C. violaceum ATCC12472 and pyocyanin, elastase and proteolytic activities, and biofilm formation of P. aeruginosa | [18] |
| 9 |  | ||||
| 10 |  | ||||
| 11 |  | Nautella sp. | C. violaceum CV026 | Inhibition of pyoyanin production of P. aeruginosa PAO1, and biofilm formation in E. coli, P. aeruginosa PAO1, and Bacillus subtilis | [19] |
| 12 |  | ||||
| 13 |  | ||||
| 14 |  | Erythrobacter sp. CUA-870 | C. violaceum CV026 | Inhibition of pyoyanin production of P. aeruginosa PAO1, and biofilm formation in E. coli, P. aeruginosa PAO1, and Bacillus subtilis | [19] |
| 15 |  | ||||
| 16 |  | ||||
| 17 |  | Dietzia maris IHBB 9296 | C. violaceum CV026 | Inhibition of pyoyanin production of P. aeruginosa PAO1, and biofilm formation in E. coli, P. aeruginosa PAO1, and Bacillus subtilis | [19] |
| 18 |  | ||||
| 19 |  | ||||
| 20 |  | Photobacterium | S. aureus 8325-4 | Inhibition of agrA and AgrA, and virulence factors α-hemolysin and Protein A in S. aureus | [20] |
| 21 |  | ||||
| 22: n = 1; R=H3C(CH2)5- 23: n = 1; R=H3C(CH2)8- 24: n = 2; R=H3C(CH2)5- |  | Chemical synthesis based on solonamide B and autoinducing peptides | P3-blaZ reporter strain | Inhibition of β-actamase activity controlled by S. aureus agr QS system | [26] |
| 25: R=H3C(CH2)5-; R1=H3C- 26: R=H3C(CH2)4-; R1=H3C- 27: R=H3C(CH2)6-; R1=H3C- 28: R=H3C(CH2)10-; R1=H3C- 29: R=H3C(CH2)5-; R1=H 30: R=H3C(CH2)5-; R1=(H3C)2CHCH2- |  | ||||
| 31: R=H3C-; R1=(H3C)2CHCH2- 32: R=(H3C)2CHCH2-; R1=H3C- 33: R=PhCH2-; R1=(H3C)2CHCH2- 34: R=(H3C)2CHCH2CH2- R1=(H3C)2CHCH2- 35: R=(H3C)2CHCH2- R1=(H3C)2CHCH2CH2- |  | ||||
| 36 |  | ||||
| 37 |  | ||||
| 38 |  | ||||
| 39 |  | ||||
40: R=  41: R=  |  | ||||
| 42: R=CH3(CH2)5CH=CH(CH2)3- 43: R=CH3(CH2)10- 44: R=CH3(CH2)5CH=CH(CH2)5- 45: R=CH3(CH2)12- |  | P. halotolerans | S. aureus lacZ reporter | Inhibition of expression of rnaIII in community-associated methicillin-resistant S. aureus USA300 | [27] |
| 46 |  | V. alginolyticus G16 | S. marcescens | Inhibition of biofilm, protease, haemolysin, lipase, prodigiosin and extracellular polysaccharide secretion in S. marcescens | [28] |
| 47 |  | R. aquimaris QSI02 | C. violaceum CV026 | Inhibition of pyocyanin production, elastase activity and biofilm formation in P. aeruginosa PA01 | [29] |
| 48 | N-acylhomoserine lactonase | Tenacibaculum sp. 20J | C. violaceum CV026 | Inhibition of SdiA in E. coli | [30] |
| 49 |  | Nocardiopsis dassonvillei subsp. dassonvillei XG-8-1 | C. violaceum CV026 and P. aeruginosa QSIS-lasI biosensors | Inhibition of C. violaceum CV026 and P. aeruginosa QSIS-lasI biosensors | [32] |
| 50 |  | ||||
| 51 |  | ||||
| 52 |  | Streptomyces sp. OUCMDZ-3436 | P. aeruginosa QSIS-lasI biosensors | Inhibition of gene expression controlled by QS in P. aeruginosa QSIS-lasI biosensors | [33] |
| 53 |  | ||||
| 54 |  | ||||
| 55 |  | ||||
| 56: n = 3; 57: n = 4; 58: n = 5; 59: n = 6; 60: n = 7; 61: n = 8; 62: n = 9; 63: n = 10; 64: n = 11; 65: n = 12 |  | Chemical synthesis based on pyrones | P. aeruginosa | Inhibition of biofilm formation of P. aeruginosa | [45] |
| 66 |  | Streptomyces sp. | C. violaceum ATCC 12472and P. aeruginosa ATCC 27853 | Inhibition of swarming, pyocyanin, biofilm formation, rhamnolipid production in P. aeruginosa | [46] |
| 67 |  | ||||
| 68 |  | Chemical synthesis based on cinnamic acid | C. violaceum | Inhibition of violacein of C. violaceum | [49] |
| 69 |  | ||||
| 70 |  | ||||
| 71 |  | ||||
| 72 |  | ||||
| 73 |  | ||||
| 74 |  | ||||
| 75 |  | Chemical synthesis based on cinnamic acid | Vibrio spp | Inhibition of biofilm formation, pigment production and protease production in Vibrio spp. | [50] |
| 76 |  | ||||
| 77 |  | ||||
| 78: R1=H; R2=C2H5- 79: R1=H; R2=C10H21- 80: R1=CH3O-; R2=C6H13- 81: R1=H; R2=C4H9- |  | ||||
| 82 |  | S. parvulus HY026 | C. violaceum ATCC 12472 | Inhibition of violaceim of C. violaceum and prodigiosin production of Serratia proteamaculans 657 | [52] |
| 83 |  | Altenaria sp. | E. coli pSB401 | Inhibition of luminescence of E. coli pSB401 | [54] |
| 84 |  | Fusarium sp. Z10 | P. aeruginosa QSIS- lasI biosensor | Inhibition of formation of biofilm, swarming motility, and the production of virulence factors in P. aeruginosa. | [55] |
| 85 |  | Penicillium sp. QF046 | C. violaceum CV026 | Inhibition of violacein of C. violaceum CV026 | [56] |
| 86 |  | P. chrysogenium | C. violaceum CV017 | Inhibition of S. aureus FabI | [57,58] |
| 87 |  | Penicillium sp. SCS-KFD08 | C. violaceum CV026 | Inhibition of violacein of C. violaceum CV026 | [59] |
| 88 |  | ||||
| 89 |  | ||||
| 90 |  | ||||
| 91 |  | ||||
| 92 |  | ||||
| 93 |  | Sarocladium | C. violaceum CV026 | Inhibition of violacein of C. violaceum CV026 | [60] |
| 94 |  | ||||
| 95 |  | ||||
| 96 |  | Fusarium | C. violaceum CV026 | Inhibition of violacein of C. violaceum CV026 | [60] |
| 97 |  | ||||
| 98 |  | ||||
| 99: R=CH3- 100: R=H |  | ||||
| 101 |  | ||||
| 102 |  | Epicoccum | C. violaceum CV026 | Inhibition of violacein of C. violaceum CV026 | [60] |
| 103 |  | ||||
| 104 |  | ||||
| 105 |  | ||||
| 106 |  | Khuskia | C. violaceum CV026 | Inhibition of violacein of C. violaceum CV026 | [60] |
| 107 |  | ||||
| 108 |  | ||||
109: R1=  ; R2=CH3O- ; R2=CH3O-110: R1=  ; R2=CH3CH2O- ; R2=CH3CH2O-111: R1=CH3CH2CH2-; R2=CH3O- 112: R1=  ; R2=CH3O- ; R2=CH3O-113: R1=  ; R2=CH3O- ; R2=CH3O-114: R1=CH3CH2CH2CH2- R2=CH3CH2O- 115: R1=CH3CH2CH2CH2-; R2=HO- 116: R1=Br-; R2=CH3O- |  | Chemical synthesis based on fusaric acid | P. aeruginosa and Vibrio fischeri | Inhibition of the las and rhl QS system in P. aeruginosa and the lux QS system in Vibrio fischeri | [66] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Wang, B.; Lu, Y.; Guo, Y.; Sun, J.; Wei, B.; Zhang, H.; Wang, H. Quorum Sensing Inhibitors from Marine Microorganisms and Their Synthetic Derivatives. Mar. Drugs 2019, 17, 80. https://doi.org/10.3390/md17020080
Chen J, Wang B, Lu Y, Guo Y, Sun J, Wei B, Zhang H, Wang H. Quorum Sensing Inhibitors from Marine Microorganisms and Their Synthetic Derivatives. Marine Drugs. 2019; 17(2):80. https://doi.org/10.3390/md17020080
Chicago/Turabian StyleChen, Jianwei, Bixia Wang, Yaojia Lu, Yuqi Guo, Jiadong Sun, Bin Wei, Huawei Zhang, and Hong Wang. 2019. "Quorum Sensing Inhibitors from Marine Microorganisms and Their Synthetic Derivatives" Marine Drugs 17, no. 2: 80. https://doi.org/10.3390/md17020080