Characterization of the Microbial Population Inhabiting a Solar Saltern Pond of the Odiel Marshlands (SW Spain)
Abstract
:1. Introduction
2. Results
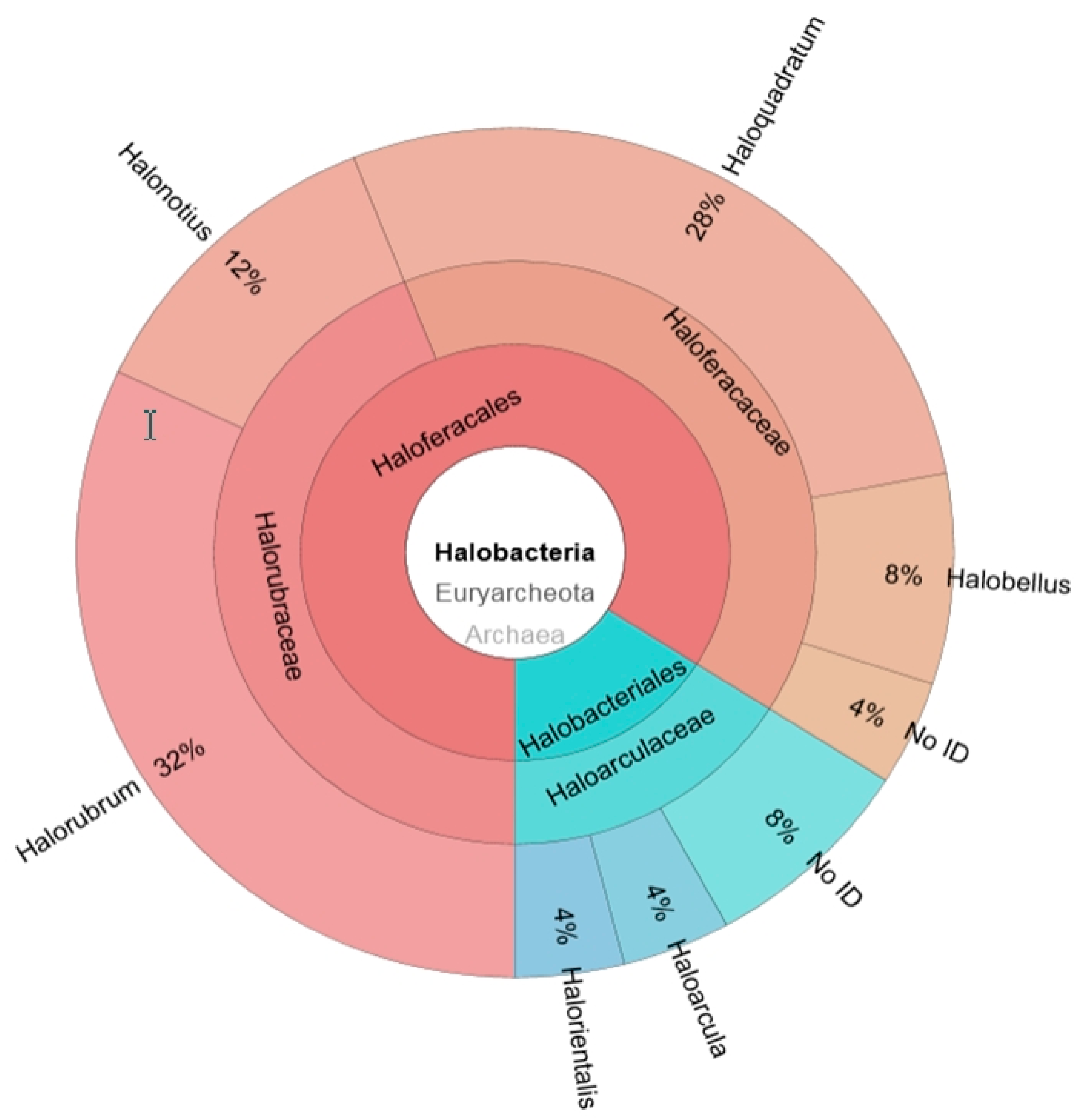
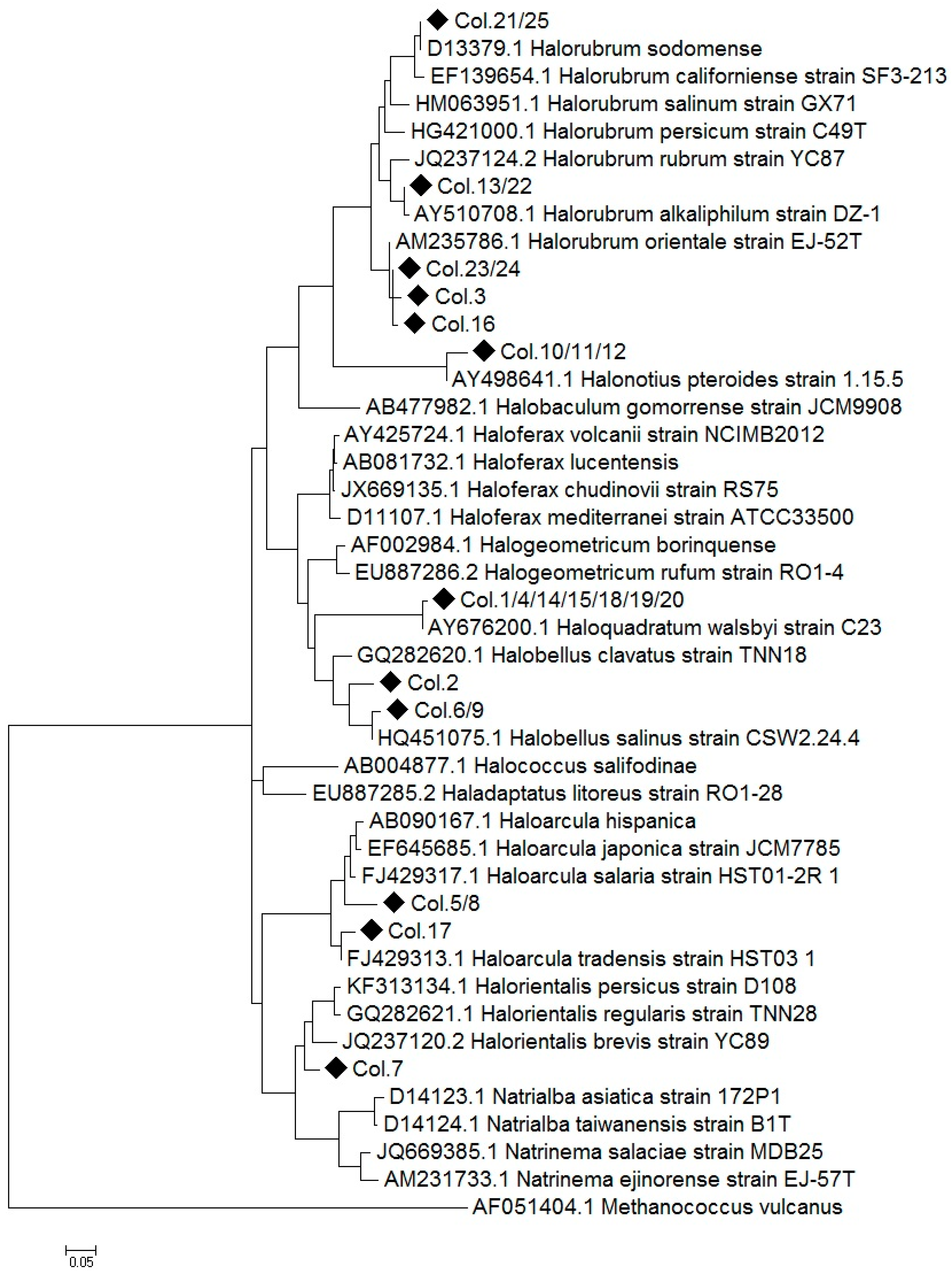
2.1. Construction of a 16S rRNA Library and Identification of the Obtained Sequences
2.2. Metagenomic Microbial Profiling by High-throughput 16S rRNA Sequencing
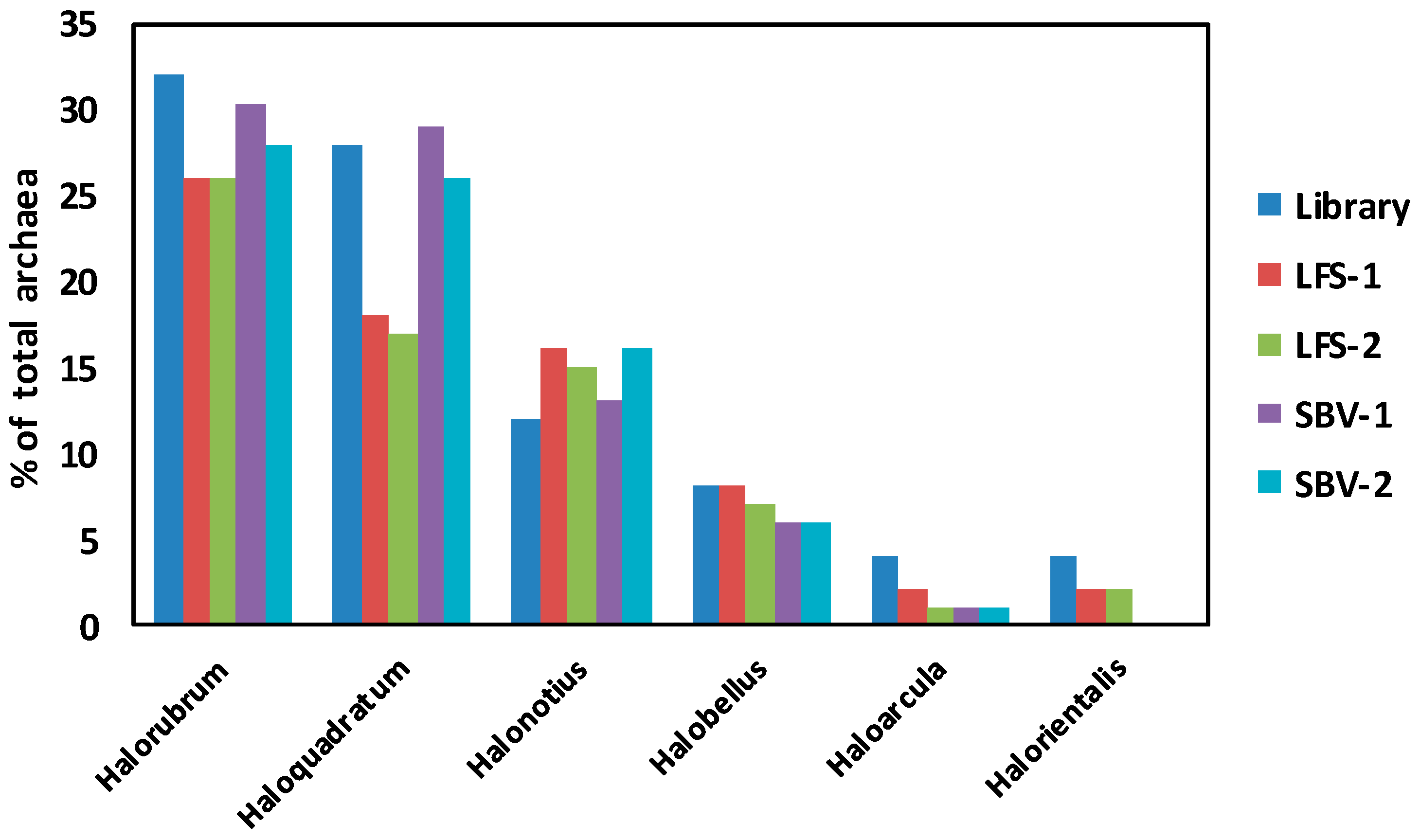
2.3. Comparison of Clone Library and 16S rRNA Metagenomic Approaches to Identify the Archaeal Microbiota of the Odiel Saltern Ponds Water
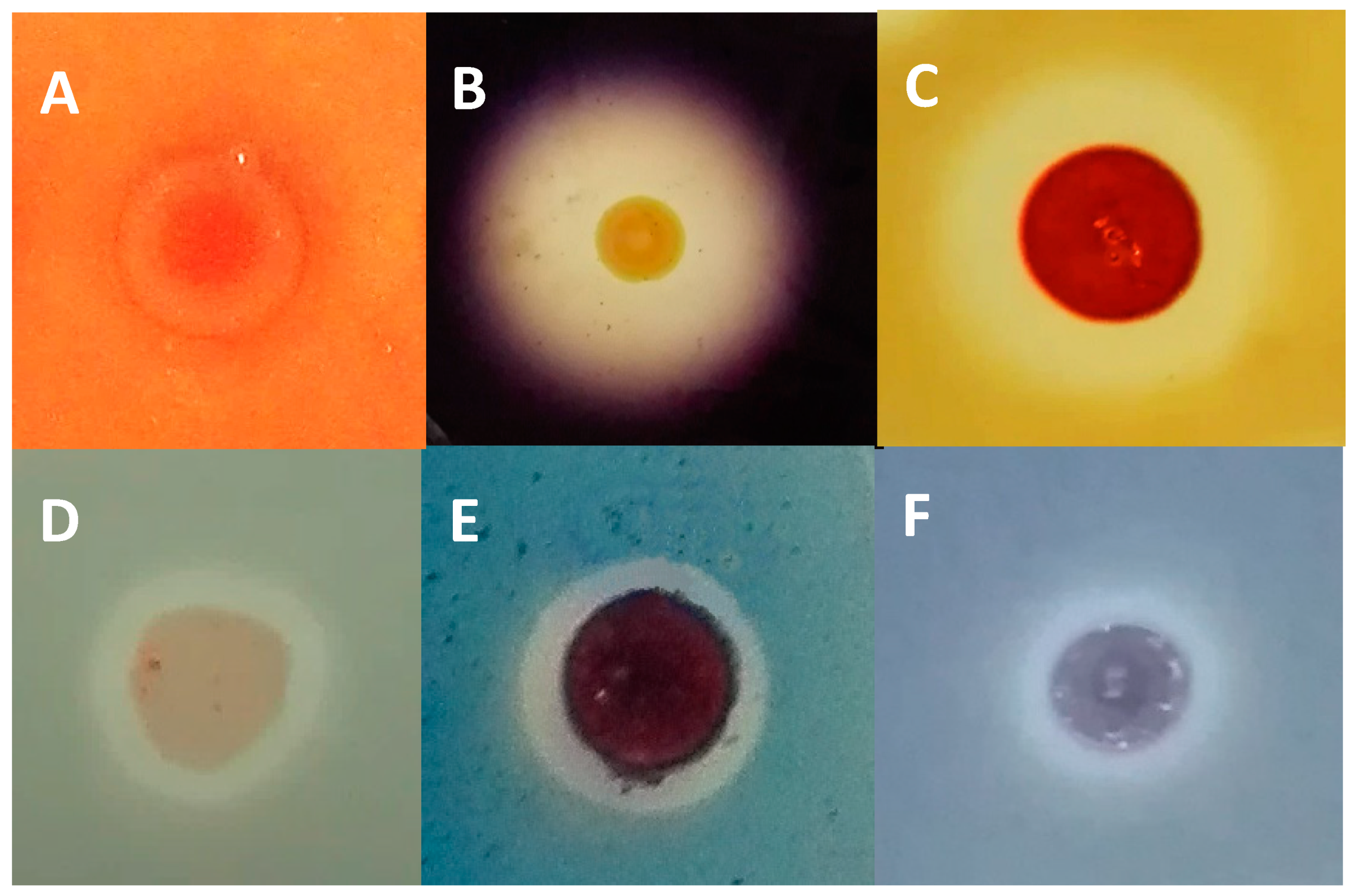
2.4. Evaluation of Halocin Activity
2.5. Haloenzymes Production by the Archaeal Enriched Biomass Isolated from the Odiel Saltern Ponds
3. Discussion
3.1. Microbiological Diversity in Hypersaline Solar Saltern Ponds
3.2. PCR Library versus 16S rRNA Massive Sequencing
3.3. Archaeal Halo-Exoenzymes
4. Materials and Methods
4.1. Sample Collection and Chemical Composition of the Brine
4.2. Genomic DNA Extraction
4.3. Amplification of 16S rRNA Encoding Gene and Construction of Clone Libraries
4.4. Construction and Analysis of Clone Libraries
4.5. High-Throughput 16S rRNA Sequencing
4.6. Extracellular Hydrolases Test
4.7. Growth Inhibition Test
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oren, A. Halophilic microbial communities and their environments. Curr. Opin. Biotechnol. 2015, 33, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Zaremba-Niedzwiedzka, K.; Caceres, E.F.; Saw, J.H.; Backstrom, D.; Juzokaite, L.; Vancaester, E.; Seitz, K.W.; Anantharaman, K.; Starnawski, P.; Kjeldsen, K.U.; et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017, 541, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, C.D. Potential for industrial products from the halophilic Archaea. J. Ind. Microbiol. Biotechnol. 2011, 38, 1635–1647. [Google Scholar] [CrossRef] [PubMed]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F1000Research 2016, 5, 396. [Google Scholar] [CrossRef] [PubMed]
- Mandelli, F.; Miranda, V.S.; Rodrigues, E.; Mercadante, A.Z. Identification of carotenoids with high antioxidant capacity produced by extremophile microorganisms. World J. Microbiol. Biotechnol. 2012, 28, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- de la Vega, M.; Sayago, A.; Ariza, J.; Barneto, A.G.; Leon, R. Characterization of a bacterioruberin-producing Haloarchaea isolated from the marshlands of the Odiel river in the southwest of Spain. Biotechnol. Prog. 2016, 32, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Karan, R.; Kapoor, S.; Singh, S.P.; Khare, S.K. Screening and isolation of halophilic bacteria producing industrially important enzymes. Brazilian J. Microbiol. 2012, 43, 1595–1603. [Google Scholar] [CrossRef] [Green Version]
- Price, L.B.; Shand, R.F. Halocin S8: A 36-amino-acid microhalocin from the haloarchaeal strain S8a. J. Bacteriol. 2000, 182, 4951–4958. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, G.; Bu, T.; Zhang, Y.; Wang, Y.; Liu, M.; Lin, X. Phylogenetic analysis and screening of antimicrobial and cytotoxic activities of moderately halophilic bacteria isolated from the Weihai Solar Saltern (China). World J. Microbiol. Biotechnol. 2010, 26, 879–888. [Google Scholar] [CrossRef]
- Kumar, S.; Grewal, J.; Sadaf, A.; Hemamalini, R.; Khare, S.K. Halophiles as a source of polyextremophilic α-amylase for industrial applications. AIMS Microbiol. 2016, 2, 1–26. [Google Scholar] [CrossRef]
- Oren, A. Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 2010, 31, 825–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thombre, R.S.; Shinde, V.D.; Oke, R.S.; Dhar, S.K.; Shouche, Y.S. Biology and survival of extremely halophilic archaeon Haloarcula marismortui RR12 isolated from Mumbai salterns, India in response to salinity stress. Sci. Rep. 2016, 6, 25642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolhuis, H.; Te Poele, E.M.; Rodriguez-Valera, F. Isolation and cultivation of Walsby’s square archaeon. Environ. Microbiol. 2004, 6, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Murcia, A.J.; Acinas, S.G.; Rodriguez-Valera, F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol. Ecol. 1995, 17, 247–255. [Google Scholar] [CrossRef]
- Anton, J.; Llobet-Brossa, E.; Rodriguez-Valera, F.; Amann, R. Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ. Microbiol. 1999, 1, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Benlloch, S.; López-López, A.; Casamayor, E.O.; Øvreås, L.; Goddard, V.; Daae, F.L.; Smerdon, G.; Massana, R.; Joint, I.; Thingstad, F.; et al. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 2002, 4, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Narasingarao, P.; Podell, S.; Ugalde, J.A.; Brochier-Armanet, C.; Emerson, J.B.; Brocks, J.J.; Heidelberg, K.B.; Banfield, J.F.; Allen, E.E. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. Isme J. 2011, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.B.; Vera-Gargallo, B.; Sánchez-Porro, C.; Ghai, R.; Papke, R.T.; Rodriguez-Valera, F.; Ventosa, A. Comparison of prokaryotic community structure from Mediterranean and Atlantic saltern concentrator ponds by a metagenomic approach. Front. Microbiol. 2014, 5, 196. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F.; Pace, N.R. Environmental Diversity of Bacteria and Archaea. Syst. Biol. 2001, 50, 470–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vera-Gargallo, B.; Ventosa, A. Metagenomic Insights into the Phylogenetic and Metabolic Diversity of the Prokaryotic Community Dwelling in Hypersaline Soils from the Odiel Saltmarshes (SW Spain). Genes 2018, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.G.; Camakaris, H.M.; Janssen, P.H.; Dyall-Smith, M.L. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 2004, 70, 5258–5265. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Thomas, R.H. Molecular Evolution and Phylogenetics. Heredity 2001, 86, 385. [Google Scholar] [CrossRef]
- Antón, J.; Rosselló-mora, R.; Amann, R.; Anto, J. Extremely Halophilic Bacteria in Crystallizer Ponds from Solar Salterns Extremely Halophilic Bacteria in Crystallizer Ponds from Solar Salterns. Appl. Environ. Microbiol. 2000, 66, 3052–3057. [Google Scholar] [CrossRef] [PubMed]
- Keshri, J.; Mishra, A.; Jha, B. Microbial population index and community structure in saline-alkaline soil using gene targeted metagenomics. Microbiol. Res. 2013, 168, 165–173. [Google Scholar] [CrossRef] [PubMed]
- León, M.J.; Aldeguer-Riquelme, B.; Antón, J.; Sánchez-Porro, C.; Ventosa, A. Spiribacter aquaticus sp. nov., a novel member of the genus Spiribacter isolated from a saltern. Int. J. Syst. Evol. Microbiol. 2017, 67, 2947–2952. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Hallsworth, J.E. Microbial weeds in hypersaline habitats: The enigma of the weed-like Haloferax mediterranei. FEMS Microbiol. Lett. 2014, 359, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Porter, K.; Russ, B.; Burns, D.; Dyall-Smith, M. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 2010, 14, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Dillon, J.G.; Carlin, M.; Gutierrez, A.; Nguyen, V.; McLain, N. Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern, Baja CA Sur, Mexico. Front. Microbiol. 2013, 4, 399. [Google Scholar] [CrossRef] [PubMed]
- Ventosa, A.; Fernández, A.B.; León, M.J.; Sánchez-Porro, C.; Rodriguez-Valera, F. The Santa Pola saltern as a model for studying the microbiota of hypersaline environments. Extremophiles 2014, 18, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Cray, J.A.; Bell, A.N.W.; Bhaganna, P.; Mswaka, A.Y.; Timson, D.J.; Hallsworth, J.E. The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 2013, 6, 453–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çınar, S.; Mutlu, M.B. Comparative analysis of prokaryotic diversity in solar salterns in eastern Anatolia (Turkey). Extremophiles 2016, 20, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Maturrano, L.; Santos, F.; Rosselló-Mora, R.; Antón, J. Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl. Environ. Microbiol. 2006, 72, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Pašić, L.; Bartual, S.G.; Ulrih, N.P.; Grabnar, M.; Velikonja, B.H. Diversity of halophilic archaea in the crystallizers of an Adriatic solar saltern. FEMS Microbiol. Ecol. 2005, 54, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Kambourova, M.; Tomova, I.; Boyadzhieva, I.; Radchenkova, N.; Vasileva-Tonkova, E. Unusually High Archaeal Diversity in a Crystallizer Pond, Pomorie Salterns, Bulgaria, Revealed by Phylogenetic Analysis. Archaea 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Payá, G.; Bautista, V.; Camacho, M.; Castejón-Fernández, N.; Alcaraz, L.A.; Bonete, M.J.; Esclapez, J. Small RNAs of Haloferax mediterranei: Identification and potential involvement in nitrogen metabolism. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.S.; Pietilä, M.K.; Oksanen, H.M. Diverse antimicrobial interactions of halophilic archaea and bacteria extend over geographical distances and cross the domain barrier. Microbiologyopen 2013, 2, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, J.C.; Burns, B.P. Untapped resources: Biotechnological potential of peptides and secondary metabolites in archaea. Archaea 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibtan, A.; Park, K.; Woo, M.; Shin, J.K.; Lee, D.W.; Sohn, J.H.; Song, M.; Roh, S.W.; Lee, S.J.; Lee, H.S. Diversity of extremely halophilic archaeal and bacterial communities from commercial salts. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pimentel, J.L.; Miller, A.Z.; Jurado, V.; Laiz, L.; Pereira, M.F.C.; Saiz-Jimenez, C. Yellow coloured mats from lava tubes of La Palma (Canary Islands, Spain) are dominated by metabolically active Actinobacteria. Sci. Rep. 2018, 8, 1944. [Google Scholar] [CrossRef] [PubMed]
- Magurran, A.E. Measuring Biological Diversity; John Wiley & Sons, 2013; ISBN 1118687922. [Google Scholar]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amoozegar, M.A.; Siroosi, M.; Atashgahi, S.; Smidt, H.; Ventosa, A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiol. 2017, 163, 623–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreck, S.D.; Grunden, A.M. Biotechnological applications of halophilic lipases and thioesterases. Appl. Microbiol. Biotechnol. 2014, 98, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Gunny, A.A.N.; Arbain, D.; Edwin Gumba, R.; Jong, B.C.; Jamal, P. Potential halophilic cellulases for in situ enzymatic saccharification of ionic liquids pretreated lignocelluloses. Bioresour. Technol. 2014, 155, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, S.; Tahmasbi, H.; Mogharabi, M.; Firuzyar, S.; Ameri, A.; Khoshayand, M.R.; Faramarzi, M.A. Efficient decolorization and detoxification of reactive orange 7 using laccase isolated from paraconiothyrium variabile, kinetics and energetics. J. Taiwan Inst. Chem. Eng. 2015, 56, 113–121. [Google Scholar] [CrossRef]
- Vithanage, L.N.G.; Barbosa, A.M.; Borsato, D.; Dekker, R.F.H. Value adding of poplar hemicellulosic prehydrolyzates: Laccase production by Botryosphaeria rhodina MAMB-05 and its application in the detoxification of prehydrolyzates. BioEnergy Res. 2015, 8, 657–674. [Google Scholar] [CrossRef]
- Waditee-Sirisattha, R.; Kageyama, H.; Takabe, T. Halophilic microorganism resources and their applications in industrial and environmental biotechnology. AIMS Microbiol. 2016, 2, 42–54. [Google Scholar] [CrossRef]
- Bajpai, B.; Chaudhary, M.; Saxena, J. Production and Characterization of alpha-Amylase from an Extremely Halophilic Archaeon, Haloferax sp. HA10. Food Technol. Biotechnol. 2015, 53, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Santorelli, M.; Maurelli, L.; Pocsfalvi, G.; Fiume, I.; Squillaci, G.; La Cara, F.; Del Monaco, G.; Morana, A. Isolation and characterisation of a novel alpha-amylase from the extreme haloarchaeon Haloterrigena turkmenica. Int. J. Biol. Macromol. 2016, 92, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Uzyol, K.S.; Akbulut, B.S.; Denizci, A.A.; Kazan, D. Thermostable a-amylase from moderately halophilic Halomonas sp. AAD21. Turk. J. Biol. 2012, 36, 327–338. [Google Scholar] [CrossRef]
- Camacho, R.M.; Mateos, J.C.; Gonzalez-Reynoso, O.; Prado, L.A.; Cordova, J. Production and characterization of esterase and lipase from Haloarcula marismortui. J. Ind. Microbiol. Biotechnol. 2009, 36, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Akolkar, A.V.; Deshpande, G.M.; Raval, K.N.; Durai, D.; Nerurkar, A.S.; Desai, A.J. Organic solvent tolerance of Halobacterium sp. SP1 (1) and its extracellular protease. J. Basic Microbiol. 2008, 48, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, D.M.; De Castro, R.E. Effect of organic solvents on the activity and stability of an extracellular protease secreted by the haloalkaliphilic archaeon Natrialba magadii. J. Ind. Microbiol. Biotechnol. 2007, 34, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Y.; Hsieh, Y.-R.; Ng, C.-C.; Chan, H.; Lin, H.-T.; Tzeng, W.-S.; Shyu, Y.-T. Purification and characterization of a novel halostable cellulase from Salinivibrio sp. strain NTU-05. Enzyme Microb. Technol. 2009, 44, 373–379. [Google Scholar] [CrossRef]
- Simankova, M.V.; Chernych, N.A.; Osipov, G.A.; Zavarzin, G.A. Halocella cellulolytica gen. nov., sp. nov., a new obligately anaerobic, halophilic, cellulolytic bacterium. Syst. Appl. Microbiol. 1993, 16, 385–389. [Google Scholar] [CrossRef]
- Yu, H.-Y.; Li, X. Alkali-stable cellulase from a halophilic isolate, Gracilibacillus sp. SK1 and its application in lignocellulosic saccharification for ethanol production. Biomass Bioenergy 2015, 81, 19–25. [Google Scholar] [CrossRef]
- Uthandi, S.; Saad, B.; Humbard, M.A.; Maupin-Furlow, J.A. LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl. Environ. Microbiol. 2010, 76, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, R.; Rezaei, S.; Jafari, N.; Forootanfar, H.; Khoshayand, M.R.; Faramarzi, M.A. Delignification and detoxification of peanut shell bio-waste using an extremely halophilic laccase from an Aquisalibacillus elongatus isolate. Extremophiles 2017, 21, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Gantner, S.; Andersson, A.F.; Alonso-Sáez, L.; Bertilsson, S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 2011, 84, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [PubMed]
- Teske, A.; Wawer, C.; Muyzer, G.; Ramsing, N.B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 1996, 62, 1405–1415. [Google Scholar] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 10. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sazci, A.; Erenler, K.; Radford, A. Detection of cellulolytic fungi by using Congo red as an indicator: A comparative study with the dinitrosalicyclic acid reagent method. J. Appl. Bacteriol. 1986, 61, 559–562. [Google Scholar] [CrossRef]
- Tekere, M.; Mswaka, A.Y.; Zvauya, R.; Read, J.S. Growth, dye degradation and ligninolytic activity studies on Zimbabwean white rot fungi. Enzym. Microb. Technol. 2001, 28, 420–426. [Google Scholar] [CrossRef]
- Lanka, S.; Latha, J.N.L. A short review on various screening methods to isolate potential lipase producers: Lipases-the present and future enzymes of biotech industry. Int. J. Biol. Chem. 2015, 9, 207–219. [Google Scholar] [CrossRef]
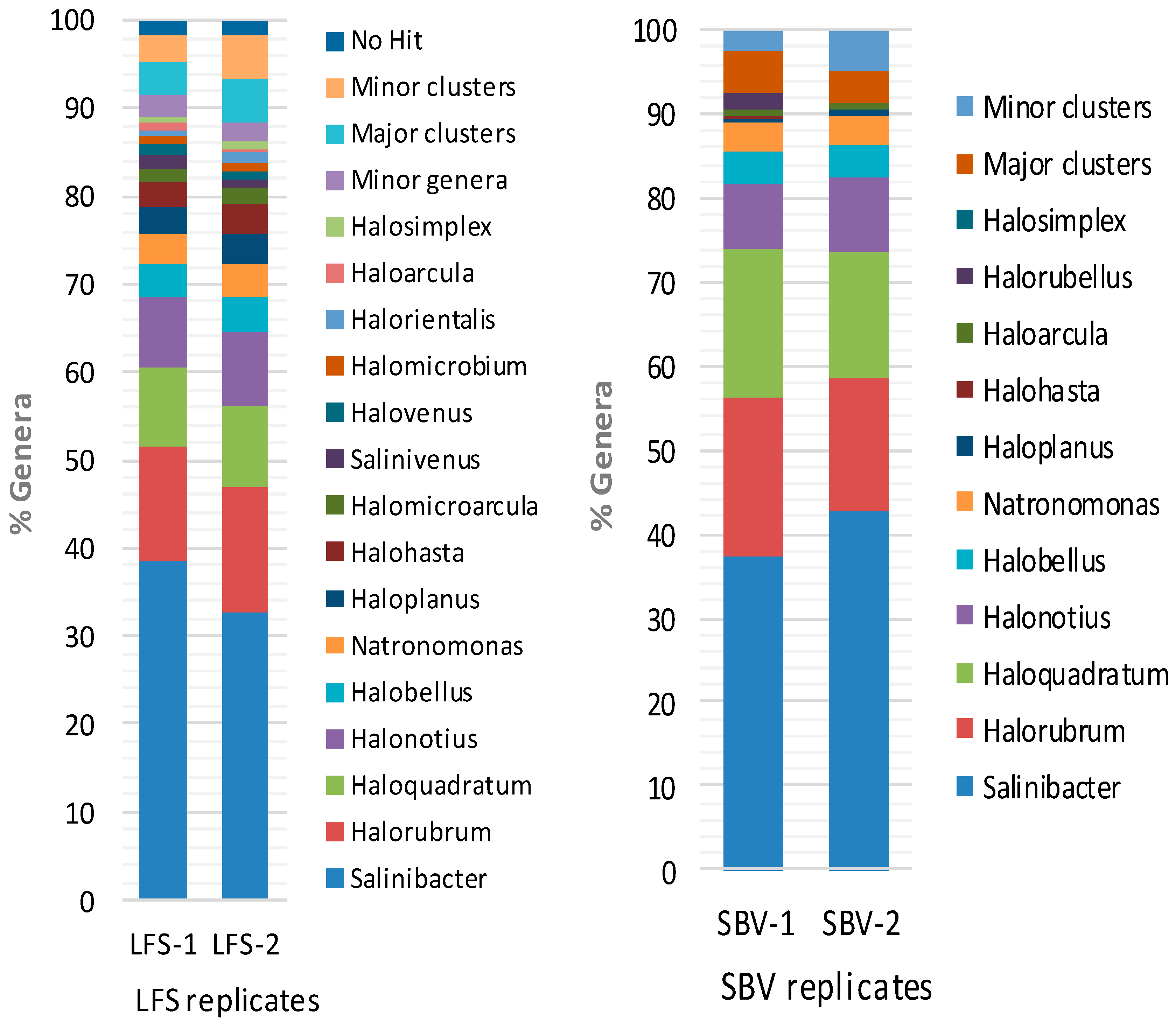
| Density (g·mL−1) | Brine Composition (g·L−1) | Total Salinity | |||||
|---|---|---|---|---|---|---|---|
| CaSO4 | MgSO4 | MgCl2 | NaCl | KCl | NaBr | ||
| 1.212 | 1.40 | 23.06 | 34.08 | 265.38 | 7.51 | 0.84 | 332.30 |
| Reaction | Raw Sequence Reads | Mean Read Length (bp) | Sequences after Denoising | Mean Quality (Q Score) | OTUs | Shannon Index |
|---|---|---|---|---|---|---|
| SBV-1 | 349 726 | 250 | 49 100 | >28 | 177 | 2.75 |
| SBV-2 | 204 766 | 250 | 19 537 | >28 | 117 | 2.65 |
| LFS-1 | 57 148 | 299.8 | 25 479 | 37.16 | 228 | 2.77 |
| LFS-2 | 156 520 | 299.6 | 71 623 | 37.25 | 356 | 3.03 |
| Sample | Santa Pola (Spain) | Santa Pola (Spain) | Odiel Salterns (Spain) * | Pomorie (Bulgaria) ** | Bajool (Australia) ** |
|---|---|---|---|---|---|
| Salinity | 33% | 37% | 33% | 34% | 34% |
| Ref. | [18,30] | [30] | This study | [35] | [28] |
| % | Haloquadratum 29.5 Haloruburm 23.1 Natronomonas 5.7 Salinibacter 4.7 Haloplanus 3.4 | Haloquadratum 58 Salinibacter 9.1 Nanosalina 4.0 Haloruburm 3.2 Nanosalinarum 1.7 Halomicrobium 1 | Salinibacter 37.8 Halorubrum 15.5 Haloquadratum 12.8 Halonotius 8.3 Halobellus 3.8 Natromonas 3.4 Haloplanus 2 Halohasta 1.7 Halorientalis 0.96 Haloarcula 0.67 | Halanaeroarchaeum 27.8 Halorubrum 24 Halonotius 15.7 Halobellus 6.5 Halovenus 6.5 Natronomonas 2.8 | Haloquadratum 47 Halorubrum 17.6 Halonotius 11.7 Haloplanus-like 11.7 Natronomonas 2.9 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Villegas, P.; Vigara, J.; León, R. Characterization of the Microbial Population Inhabiting a Solar Saltern Pond of the Odiel Marshlands (SW Spain). Mar. Drugs 2018, 16, 332. https://doi.org/10.3390/md16090332
Gómez-Villegas P, Vigara J, León R. Characterization of the Microbial Population Inhabiting a Solar Saltern Pond of the Odiel Marshlands (SW Spain). Marine Drugs. 2018; 16(9):332. https://doi.org/10.3390/md16090332
Chicago/Turabian StyleGómez-Villegas, Patricia, Javier Vigara, and Rosa León. 2018. "Characterization of the Microbial Population Inhabiting a Solar Saltern Pond of the Odiel Marshlands (SW Spain)" Marine Drugs 16, no. 9: 332. https://doi.org/10.3390/md16090332





