Gageostatins A–C, Antimicrobial Linear Lipopeptides from a Marine Bacillus subtilis
Abstract
:1. Introduction
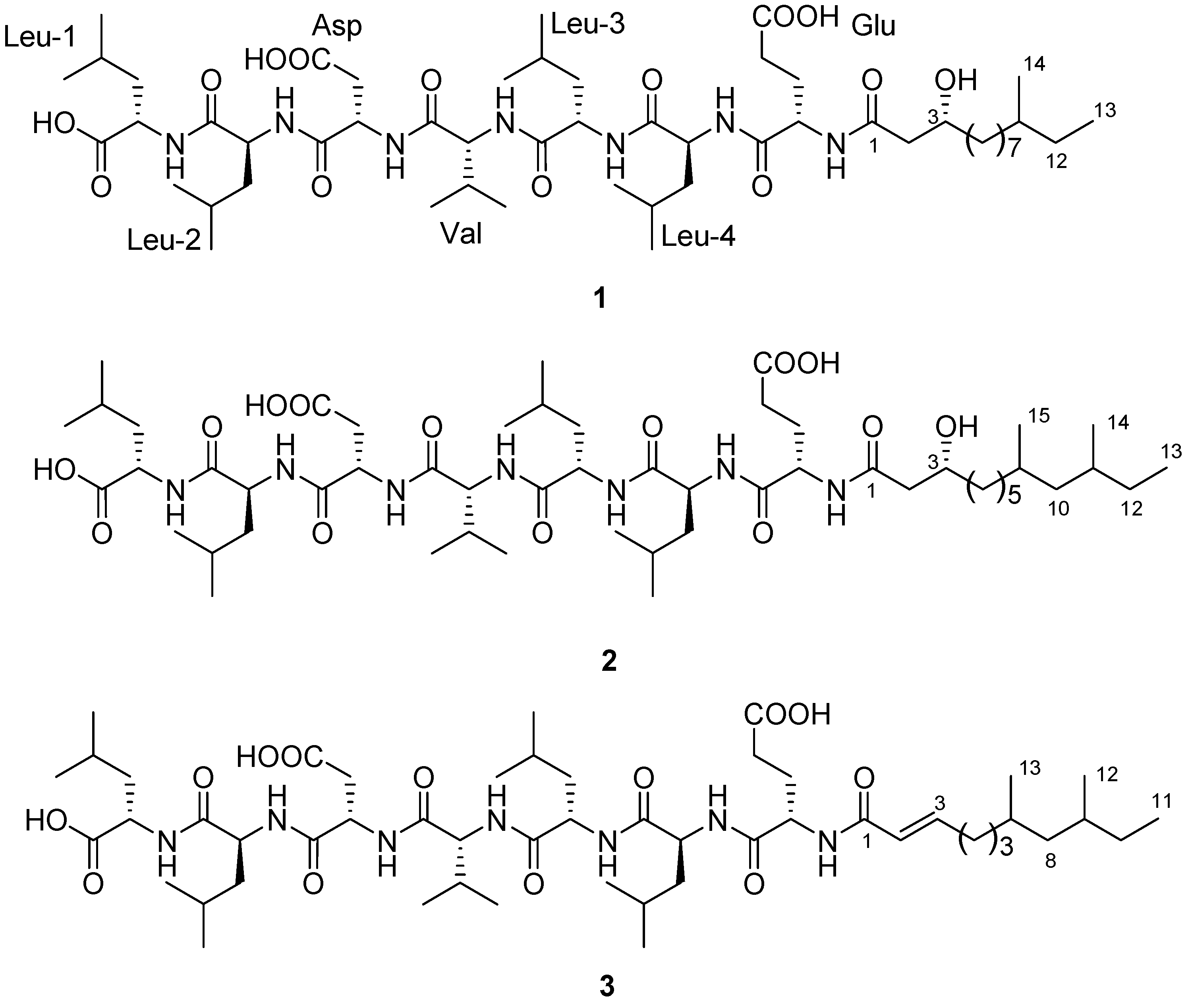
2. Results and Discussion
2.1. Isolation of Compounds
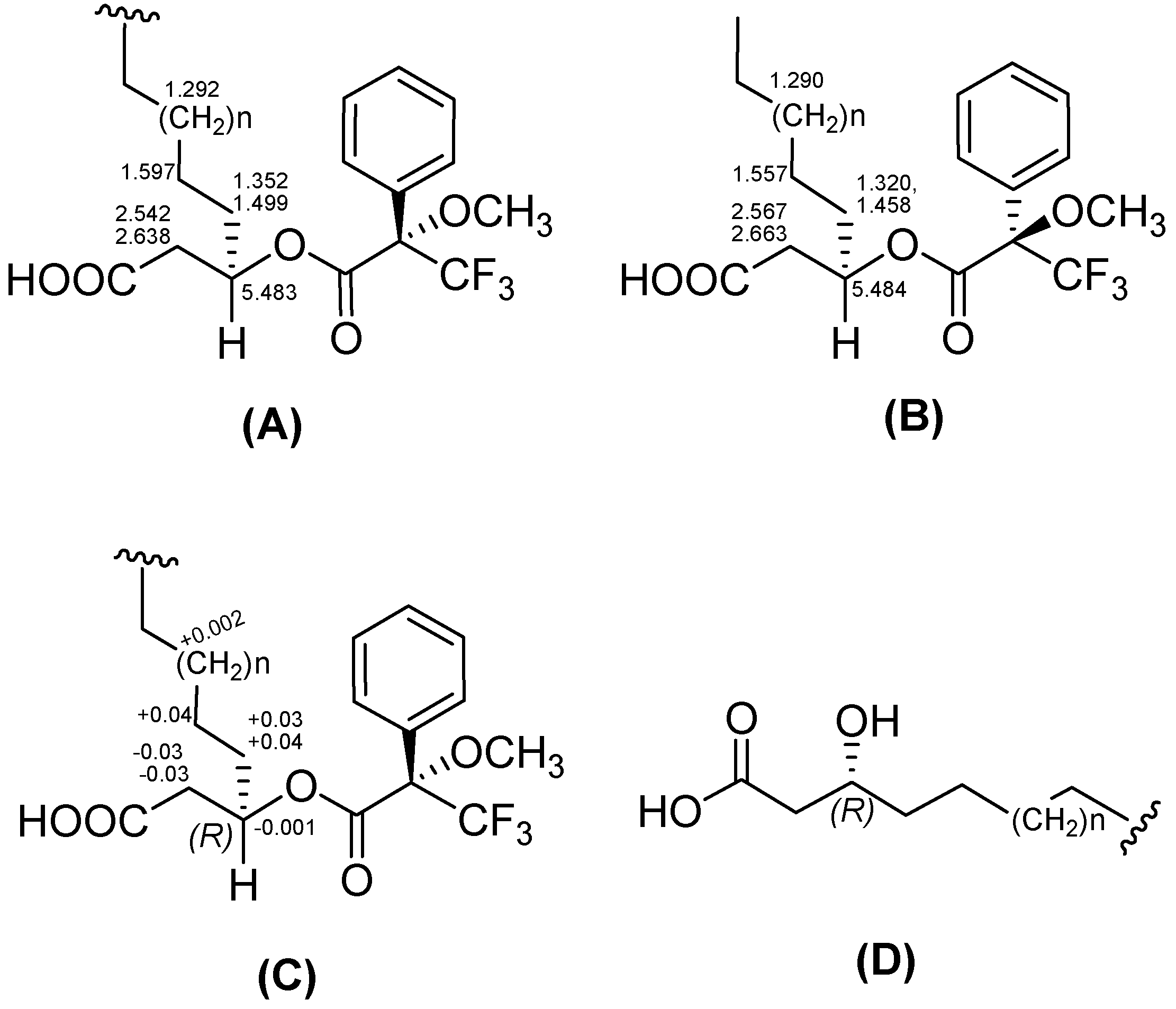
2.2. Structure Determination
| 1 | 2 | 3 | |||||
|---|---|---|---|---|---|---|---|
| Units | No. | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) | δC |
| Leu-1 | |||||||
| 1 | 180.3 | 180.2 | 180.2 | ||||
| 2 | 4.37 m | 53.5 | 4.37 m | 53.3 | 4.30 m | 54.8 | |
| 3 | 1.65 m | 41.6 | 1.65 m | 41.6 | 1.63 m | 41.6 | |
| 4 | 1.65 m | 26.1 | 1.65 m | 26.1 | 1.63 m | 26.1 | |
| 5 | 0.91 m | 21.7 | 0.91 m | 21.7 | 0.89 m | 21.8 | |
| 6 | 0.91 m | 23.5 | 0.91 m | 23.4 | 0.89 m | 23.4 | |
| NH | 8.37 d (8.0) a | 8.37 d (8.0) a | 8.52 d (9.5) a | ||||
| Leu-2 | |||||||
| 1 | 173.6 | 173.5 | 175.7 | ||||
| 2 | 4.38 m | 53.4 | 4.38 m | 53.3 | 4.39 m | 53.3 | |
| 3 | 1.65 m | 43.4 | 1.65 m | 43.4 | 1.63 m | 41.4 | |
| 4 | 1.65 m | 26.1 | 1.65 m | 26.1 | 1.63 m | 26.1 | |
| 5 | 0.91 m | 21.7 | 0.91 m | 21.7 | 0.96 m | 22.3 | |
| 6 | 0.91 m | 23.6 | 0.91 m | 23.6 | 0.96 m | 24.0 | |
| NH | 7.96 d (8.0) a | 7.97 d (8.0) a | 7.67 d (9.0) a | ||||
| Asp | |||||||
| 1 | 174.3 | 174.2 | 174.3 | ||||
| 2 | 4.57 t (6.0) | 53.1 | 4.57 m | 53.0 | 4.56 m | 53.2 | |
| 3 | 2.56 dd (16.5, 5.5) | 40.1 | 2.56 dd (16.5, 5.0) | 40.0 | 2.51 dd (16.5, 5.5) | 40.1 | |
| 2.76 dd (16.5, 6.0) | 2.75 dd (16.5, 5.5) | 2.75 dd (16.5, 6.0) | |||||
| NH | 8.38 d (8.0) a | 8.39 d (8.0) a | 8.40 d (9.0) a | ||||
| COOH | 178.2 | 178.0 | 178.2 | ||||
| Val | |||||||
| 1 | 173.5 | 173.5 | 173.7 | ||||
| 2 | 4.13 d (8.0) | 61.0 | 4.13 m | 60.9 | 4.06 m | 61.2 | |
| 3 | 2.18 m | 31.2 | 2.17 m | 31.2 | 2.13 m | 31.2 | |
| 4 | 0.93 m | 19.3 | 0.93 m | 19.7 | 0.96 m | 19.8 | |
| 5 | 0.93 m | 20.2 | 0.93 m | 20.0 | 0.96 m | 20.1 | |
| NH | 7.96 d (8.0) a | 7.97 d (8.0) a | 7.97 d (8.0) a | ||||
| Leu-3 | |||||||
| 1 | 175.0 | 174.9 | 175.5 | ||||
| 2 | 4.40 m | 54.8 | 4.40 m | 54.7 | 4.35 m | 53.5 | |
| 3 | 1.65 m | 41.4 | 1.65 m | 41.4 | 1.65 m | 40.4 | |
| 4 | 1.65 m | 26.1 | 1.65 m | 26.1 | 1.63 m | 26.1 | |
| 5 | 0.93 m | 22.2 | 0.93 m | 22.1 | 0.92 m | 22.4 | |
| 6 | 0.93 m | 23.9 | 0.93 m | 23.8 | 0.92 m | 23.8 | |
| NH | 7.68 d (9.0) a | 7.68 d (9.0) a | 7.96 d (8.0) a | ||||
| Leu-4 | |||||||
| 1 | 175.6 | 175.5 | 173.6 | ||||
| 2 | 4.31 m | 54.2 | 4.31 m | 54.1 | 4.33 m | 55.5 | |
| 3 | 1.65 m | 41.3 | 1.65 m | 41.3 | 1.63 m | 40.4 | |
| 4 | 1.65 m | 26.1 | 1.65 m | 26.1 | 1.63 m | 26.1 | |
| 5 | 0.93 m | 22.3 | 0.93 m | 22.2 | 0.92 m | 22.3 | |
| 6 | 0.93 m | 24.1 | 0.93 m | 23.9 | 0.92 m | 23.9 | |
| NH | 8.48 d (6.5) a | 8.49 d (6.5) a | 8.51 m | ||||
| Glu | |||||||
| 1 | 175.0 | 174.9 | 175.1 | ||||
| 2 | 4.34 m | 55.5 | 4.34 m | 55.4 | 4.35 m | 55.5 | |
| 3 | 1.95 m | 29.6 | 1.95 m | 29.5 | 1.95 m | 29.8 | |
| 2.05 m | 2.05 m | 2.03 m | |||||
| 4 | 2.29 t (7.0) | 35.4 | 2.29 m | 35.4 | 2.26 m | 35.5 | |
| COOH | 181.5 | 181.4 | 181.5 | ||||
| NH | 8.79 d (6.5) a | 8.81 d (6.5) a | 8.81 brd. | ||||
| 3-OH acid | |||||||
| 1 | 175.4 | 175.3 | 169.1 | ||||
| 2 | 2.33 dd (9.0, 14.5) | 45.0 | 2.33 dd (8.5, 14.0) | 44.7 | 6.01 d (15.5) | 124.7 | |
| 2.46 dd (4.0, 14.5) | 2.47 dd (4.0, 14.5) | ||||||
| 3 | 3.98 m | 70.2 | 3.98 m | 70.0 | 6.79 dt(15.5, 8.5) | 146.5 | |
| 4 | 1.34 m | 26.9 | 1.34 m | 26.9 | 2.18 m | 33.2 | |
| 1.48 m | 1.47 m | ||||||
| 5 | 1.49 m | 38.6 | 1.49 m | 38.5 | 1.45 m | 29.7 | |
| 6 | 1.29 brs. | 28.1–31.3 | 1.29 brs. | 28.1–31.3 | 1.16 m | 40.4 | |
| 7 | 1.52 m | 29.3 | |||||
| 8 | 1.17 m | 40.3 | 1.28 brds. | 30.8 | |||
| 9 | 1.54 m | 29.2 | 1.28 brds. | 35.8 | |||
| 10 | 1.17 m | 40.4 | 1.29 brs. | 30.8 | 1.12 m | 37.9 | |
| 1.28 m | |||||||
| 11 | 1.51 m | 29.3 | 1.29 brs. | 35.7 | 0.87 m | 11.9 | |
| 12 | 1.29 brs. | 38.5 | 1.12 m | 37.8 | 0.86 m | 14.6 | |
| 1.29 m | |||||||
| 13 | 0.86 m | 14.6 | 0.87 m | 11.9 | 0.87 m | 23.2 | |
| 14 | 0.87 m | 23.1 | 0.86 m | 14.5 | |||
| 15 | 0.87 m | 23.1 | |||||
 −29 (0.1, MeOH) (SI). The lipopeptide structure of 1 was then corroborated constructing the sequence of Leu-Leu-Asp-Val-Leu-Leu-Glu-fatty acid by two dimensional ROESY and HMBC experiments which showed long range proton-proton and proton-carbon correlations and correlations between amide protons and α-protons.
−29 (0.1, MeOH) (SI). The lipopeptide structure of 1 was then corroborated constructing the sequence of Leu-Leu-Asp-Val-Leu-Leu-Glu-fatty acid by two dimensional ROESY and HMBC experiments which showed long range proton-proton and proton-carbon correlations and correlations between amide protons and α-protons.  −6 (0.25, MeOH). The search result from the natural products database system suggested this fatty acid is a new fatty acid derivative. Finally, the complete chemical structure of compound 2 was constructed by the detailed analysis of COSY, TOCSY, HMBC and ROESY correlations (Supplementary Information).
−6 (0.25, MeOH). The search result from the natural products database system suggested this fatty acid is a new fatty acid derivative. Finally, the complete chemical structure of compound 2 was constructed by the detailed analysis of COSY, TOCSY, HMBC and ROESY correlations (Supplementary Information). 
2.3. Antimicrobial Activities
| MICs (µg/mL) | |||||
|---|---|---|---|---|---|
| Microorganisms | 1 | 2 | 1 + 2 | 3 | P.C. |
| Fungi | |||||
| R. solani | 4 | 8 | 4 | 32 | 1 |
| C. acutatum | 8 | 8 | 4 | 16 | 1 |
| B. cinera | 4 | 8 | 4 | 32 | 1 |
| Gram Positive Bacteria | |||||
| S. aureus | 16 | 16 | 8 | 64 | 2 |
| B. subtilis | 16 | 32 | 16 | 32 | 2 |
| Gram Negative Bacteria | |||||
| S. Typhi | 16 | 32 | 32 | 32 | 2 |
| P. aeruginosa | 16 | 16 | 8 | 64 | 2 |
2.4. Cytotoxic Properties
| Cancer Cell Lines | (GI50, µg/mL) | ||||
|---|---|---|---|---|---|
| 1 | 2 | 1 + 2 | 3 | ADR a | |
| Breast cancer: MDA-MB-231 | 14.9 | 16.1 | 10.5 | 11.2 | 0.56 |
| Colon cancer: HCT-15 | 11.4 | 18.3 | 10.9 | 23.2 | 0.33 |
| Prostate cancer: PC-3 | 10.8 | 19.4 | 12.0 | 11.7 | 0.91 |
| Lung cancer: NCI-H23 | 11.2 | 11.7 | 4.6 | 10.9 | 0.71 |
| Stomach cancer: NUGC-3 | 11.8 | 13.9 | 10.1 | 10.5 | 0.53 |
| Renal cancer: ACHN | 11.5 | 18.4 | 10.7 | 12.3 | 0.51 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Isolation and Identification of the Strain 109GGC020
3.3. Seed and Mass Cultures of the Strain
3.4. Extraction and Isolation of Compounds
 +52 (c 0.1, MeOH); IR (MeOH) νmax 3291 cm−1 (NH) and 1646–1737 cm−1 (CO), 2930 cm−1; 1H and 13C NMR data (CD3OD), Table 1; HRESIMS m/z 1062.6691 [M + Na]+.
+52 (c 0.1, MeOH); IR (MeOH) νmax 3291 cm−1 (NH) and 1646–1737 cm−1 (CO), 2930 cm−1; 1H and 13C NMR data (CD3OD), Table 1; HRESIMS m/z 1062.6691 [M + Na]+. +53 (c 0.1, MeOH); IR (MeOH) νmax 3293 (br), 2927, 1742 cm−1; 1H and 13C NMR data (CD3OD), Table 1; HRESIMS m/z 1076.6831 [M + Na]+.
+53 (c 0.1, MeOH); IR (MeOH) νmax 3293 (br), 2927, 1742 cm−1; 1H and 13C NMR data (CD3OD), Table 1; HRESIMS m/z 1076.6831 [M + Na]+. +16 (c 0.1, MeOH); IR (MeOH) νmax 3373 (br), 2970, 1660 cm−1; 1H and 13C NMR data (CD3OD), Table 1; HRESIMS m/z 1030.6423 [M + Na]+.
+16 (c 0.1, MeOH); IR (MeOH) νmax 3373 (br), 2970, 1660 cm−1; 1H and 13C NMR data (CD3OD), Table 1; HRESIMS m/z 1030.6423 [M + Na]+.3.5. Acid Hydrolysis of Compounds 1–3
 −29 (0.1, MeOH).
−29 (0.1, MeOH).3.6. Preparation of the (S)- and (R)-MTPA Esters of 3-Hydroxy Fatty Acids (1b and 1c)
3.7. Advanced Marfey’s Analysis of 1–3
3.8. Antimicrobial Assays
3.9. Cytotoxicity Test
4. Conclusions
Supplementary Files
Acknowledgments
Conflicts of Interest
References
- Ebada, S.S.; Lin, W.H.; Proksch, P. Review: Bioactive sesterterpenes and triterpenes from marine sponges: Occurrence and pharmacological significance. Mar. Drugs 2010, 8, 313–346. [Google Scholar] [CrossRef]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Rampelotto, P.H. Review: Resistance of microorganisms to extreme environmental conditions and its contribution to astrobiology. Sustainability 2010, 2, 1602–1623. [Google Scholar] [CrossRef]
- Earl, A.M.; Losick, R.; Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008, 16, 269–275. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.; Azevedo, V.; Bertero, M.G.; Bessières, P.; Bolotin, A.; Borchert, S. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef]
- Fan, L.; Bo, S.; Chen, H.; Ye, W.; Kleinschmidt, K.; Baumann, H.I.; Imhoff, J.F.; Kleine, M.; Cai, D. Genome sequence of Bacillus subtilis subsp. spizizenii gtP20b, isolated from the Indian ocean. J. Bacteriol. 2011, 193, 1276–1277. [Google Scholar] [CrossRef]
- Hamdache, A.; Lamarti, A.; Aleu, J.; Collado, I.G. Non-peptide metabolites from the genus Bacillus. J. Nat. Prod. 2011, 74, 893–899. [Google Scholar] [CrossRef]
- Baruzzi, F.; Quintieri, L.; Morea, M.; Caputo, L. Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Vilas, A.M., Ed.; Formatex: Badajoz, Spain, 2011; pp. 1102–1111. [Google Scholar]
- Mondol, M.A.M.; Shin, H.J.; Islam, M.T. Diversity of secondary metabolites from marine Bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef]
- Zou, A.; Liu, J.; Garamus, V.M.; Zheng, K.; Willumeit, R.; Mu, B. Interaction between the natural lipopeptide [Glu1, Asp5] Surfactin-C15 and hemoglobin in aqueous solution. Biomacromolecules 2010, 11, 593–599. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 5, 845–857. [Google Scholar] [CrossRef]
- Makovitzki, A.; Baram, J.; Shai, Y. Antimicrobial lipopolypeptides composed of palmitoyl di- and tricationic peptides: In vitro and in vivo activities, self-assembly to nanostructures, and a plausible mode of action. Biochemistry 2008, 47, 10630–10636. [Google Scholar] [CrossRef]
- Shai, Y.; Makovitzky, A.; Avrahami, D. Host defense peptides and lipopeptides: Modes of action and potential candidates for the treatment of bacterial and fungal infections. Curr. Protein Pept. Sci. 2006, 7, 479–486. [Google Scholar] [CrossRef]
- Straus, S.K.; Hancock, R.E. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 2006, 1758, 1215–1223. [Google Scholar] [CrossRef]
- Thevissen, K.; Terras, F.R.; Broekaert, W.F. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 1999, 65, 5451–5458. [Google Scholar]
- Hobden, C.; Teevan, C.; Jones, L.; O’Shea, P. Hydrophobic properties of the cell surface of Candida albicans: A role in aggregation. Microbiology 1995, 14, 1875–1881. [Google Scholar]
- Kurtz, M.B.; Douglas, C.M. Lipopeptide inhibitors of fungal glucan synthase. J. Med. Vet. Mycol. 1997, 35, 79–86. [Google Scholar] [CrossRef]
- Thomas, D.N.; Dieckmann, G.S. Antarctic sea ice—A habitat for extremophiles. Science 2002, 295, 641–644. [Google Scholar] [CrossRef]
- Liu, X.; Ashforth, E.; Ren, B.; Song, F.; Dai, H.; Liu, M.; Wang, J.; Xie, Q.; Zhang, L. Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J. Antibiot. 2010, 63, 415–422. [Google Scholar] [CrossRef]
- Marfey, P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1-5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Ohtani, I.; Kusumi, T.; Kashman, Y.; Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configuration of marine terpenoids. J. Am. Chem. Soc. 1991, 113, 4092–4096. [Google Scholar] [CrossRef]
- Freire, F.; Seco, J.M.; Emilio, Q.; Riguera, R. Determining the absolute stereochemistry of secondary/secondary diols by 1H NMR: Basis and applications. J. Org. Chem. 2005, 70, 3778–3790. [Google Scholar]
- Jenske, R.; Vetter, W. Enantioselective analysis of 2- and 3-hydroxy fatty acids in food samples. J. Agric. Food Chem. 2008, 56, 11578–11583. [Google Scholar] [CrossRef]
- Maider, P.; Francoise, P.; Jean, W. Solid-phase synthesis of surfactin, a powerful biosurfactant produced by Bacillus subtilis, and of four analogues. Int. J. Pept. Res. Ther. 2005, 11, 195–202. [Google Scholar] [CrossRef]
- Appendio, G.; Gibbons, S.; Giana, A.; Pagani, A.; Grassi, G.; Stavri, M.; Smith, E.; Rahman, M.M. Antibacterial cannabinoids from Cannabis sativa: A structure-activity study. J. Nat. Prod. 2008, 71, 1427–1430. [Google Scholar] [CrossRef]
- Oluwatuyi, M.; Kaatz, G.W.; Gibbons, S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry 2004, 65, 3249–3254. [Google Scholar] [CrossRef]
- Yu, J.Q.; Lei, J.C.; Yu, H.D.; Cai, X.; Zou, G.L. Chemical composition and antimicrobial activity of the essential oil of Scutellaria barbata. Phytochemistry 2004, 65, 881–884. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R; Scudiero, D.; Monks, A.; McMohan, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyed, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Shaligram, N.S.; Singhal, R.S. Surfactin—A review on biosynthesis, fermentation, purification and applications. Food Technol. Biotechnol. 2010, 48, 119–134. [Google Scholar]
- Batrakov, S.G.; Rodionova, T.A; Esipov, S.E.; Polyakov, N.B.; Sheichenko, V.I.; Shekhovtsova, N.V.; Lukin, S.M.; Panikov, N.S.; Nikolaev, Y.A. A novel lipopeptide, an inhibitor of bacterial adhesion, from the thermophilic and halotolerant subsurface Bacillus licheniformis strain 603. Biochim. Biophys. Acta 2003, 1634, 107–115. [Google Scholar] [CrossRef]
- Canova, S.P.; Petta, T.; Reyes, L.F.; Zucchi, T.D.; Moraes, L.A.B.; Melo, I.S. Characterization of lipopeptides from Paenibacillus sp. (IIRAC30) suppressing Rhizoctonia solani. World J. Microbiol. Biotechnol. 2010, 26, 2241–2247. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, J.Y.; Kim, S.H.; Bae, H.J.; Yi, H.; Yoon, S.H.; Koo, B.S.; Kwon, M.; Cho, J.Y.; Lee, C.H.; et al. Surfactin from Bacillus subtilis displays anti-proliferative effect via apoptosis induction, cell cycle arrest and survival signaling suppression. FEBS Lett. 2007; 581, 865–871. [Google Scholar]
- Wang, C.L.; Ng, T.B.; Yuan, F.; Liu, Z.K.; Liu, F. Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from Bacillus subtilis natto T-2. Peptides 2007, 28, 1344–1350. [Google Scholar] [CrossRef]
- Romano, A.; Vitullo, D.; Di, P.A.; Lima, G.; Lanzotti, V. Antifungal lipopeptides from Bacillus amyloliquefaciens strain BO7. J. Nat. Prod. 2011, 74, 145–151. [Google Scholar] [CrossRef]
- Kim, H.S.; Yoon, B.D.; Lee, C.H.; Suh, H.H.; Oh, H.M.; Katsuragi, T.; Tani, Y. Production and properties of a lipopeptide biosurfactant from Bacillus subtilis C9. J. Ferment. Bioeng. 1997, 84, 41–46. [Google Scholar] [CrossRef]
- Kim, P.I.; Bai, H.; Bai, D.; Chae, H.; Chung, S.; Kim, Y.; Park, R.; Chi, Y.T. Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. J. Appl. Microbiol. 2004, 97, 942–949. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tareq, F.S.; Lee, M.A.; Lee, H.-S.; Lee, J.-S.; Lee, Y.-J.; Shin, H.J. Gageostatins A–C, Antimicrobial Linear Lipopeptides from a Marine Bacillus subtilis. Mar. Drugs 2014, 12, 871-885. https://doi.org/10.3390/md12020871
Tareq FS, Lee MA, Lee H-S, Lee J-S, Lee Y-J, Shin HJ. Gageostatins A–C, Antimicrobial Linear Lipopeptides from a Marine Bacillus subtilis. Marine Drugs. 2014; 12(2):871-885. https://doi.org/10.3390/md12020871
Chicago/Turabian StyleTareq, Fakir Shahidullah, Min Ah Lee, Hyi-Seung Lee, Jong-Seok Lee, Yeon-Ju Lee, and Hee Jae Shin. 2014. "Gageostatins A–C, Antimicrobial Linear Lipopeptides from a Marine Bacillus subtilis" Marine Drugs 12, no. 2: 871-885. https://doi.org/10.3390/md12020871
APA StyleTareq, F. S., Lee, M. A., Lee, H.-S., Lee, J.-S., Lee, Y.-J., & Shin, H. J. (2014). Gageostatins A–C, Antimicrobial Linear Lipopeptides from a Marine Bacillus subtilis. Marine Drugs, 12(2), 871-885. https://doi.org/10.3390/md12020871





