Back from the Past: DNA Barcodes and Morphology Support Ablabesmyia americana Fittkau as a Valid Species (Diptera: Chironomidae)
Abstract
:1. Introduction
2. Materials and Methods
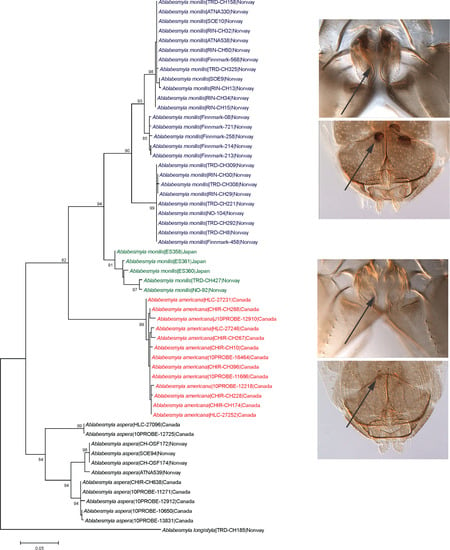
3. Results
Taxonomy
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hebert, P.; Cywinska, A.; Ball, S.; de Waard, J. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodin, Y.; Ejdung, G.; Strandberg, J.; Lyrholm, T. Improving environmental and biodiversity monitoring in the Baltic Sea using DNA barcoding of Chironomidae (Diptera). Mol. Ecol. Resour. 2012, 13, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Ekrem, T.; Willassen, E.; Stur, E. A comprehensive DNA sequence library is essential for identification with DNA barcodes. Mol. Phylogenet. Evol. 2007, 43, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Ekrem, T.; Stur, E.; Hebert, P.D.N. Females do count: Documenting Chironomidae (Diptera) species diversity using DNA barcoding. Org. Divers. Evol. 2010, 10, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Silva, F.L.D.; Ekrem, T.; Fonseca-Gessner, A.A. DNA barcodes for species delimitation in Chironomidae (Diptera): A case study on the genus Labrundinia. Can. Entomol. 2013, 145, 589–602. [Google Scholar] [CrossRef]
- Lin, X.; Stur, E.; Ekrem, T. Exploring genetic divergence in a species-rich insect genus using 2790 DNA barcodes. PLoS ONE 2015, 10, e0138993. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.M.; Stur, E.; Ekrem, T. Molecular and morphological methods reveal cryptic diversity and three new species of Nearctic Micropsectra (Diptera: Chironomidae). Freshw. Sci. 2013, 32, 892–921. [Google Scholar] [CrossRef]
- Lin, X.-L.; Stur, E.; Ekrem, T. DNA barcodes and morphology reveal unrecognized species in Chironomidae (Diptera). Insect Syst. Evol. 2018, 49, 329–398. [Google Scholar] [CrossRef] [Green Version]
- Carew, M.E.; Pettigrove, V.; Hoffmann, A.A. The utility of DNA markers in classical taxonomy: Using cytochrome oxidase I markers to differentiate Australian Cladopelma (Diptera: Chironomidae) midges. Ann. Ent. Soc. Am. 2005, 98, 587–594. [Google Scholar] [CrossRef]
- Stur, E.; Ekrem, T. Exploring unknown life stages of Arctic Tanytarsini (Diptera: Chironomidae) with DNA barcoding. Zootaxa 2011, 2743, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Silva, F.L.; Wiedenbrug, S. Integrating DNA barcodes and morphology for species delimitation in the Corynoneura group (Diptera: Chironomidae: Orthocladiinae). Bull. Ent. Res. 2014, 104, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Niitsuma, H.; Tang, H. Taxonomic review of Ablabesmyia Johannsen (Diptera: Chironomidae: Tanypodinae) from Oriental China, with descriptions of six new species. Zootaxa 2019, 4564, 248–270. [Google Scholar] [CrossRef]
- Oliveira, C.S.N.D.; Silva, M.A.N.D.; Gessner, A.A.F. Neotropical Ablabesmyia Johannsen (Diptera: Chironomidae, Tanypodinae)–Part I. Zootaxa 2013, 3733, 1–123. [Google Scholar] [CrossRef] [PubMed]
- Fusari, L.M.; Oliveira, C.S.N.; Hamada, N.; Roque, F.O. New species of Ablabesmyia Johannsen from the Neotropical region: First report of a sponge-dwelling Tanypodinae. Zootaxa 2012, 3239, 43–50. [Google Scholar] [CrossRef]
- Johannsen, O.A. Aquatic nematocerous Diptera II. Chironomidae. Bull. N. Y. State Mus. 1905, 86, 77–325. [Google Scholar]
- Edwards, F.W. British non-biting midges (Diptera, Chironomidae). Trans. Entomol. Soc. Lond. 1929, 77, 279–430. [Google Scholar] [CrossRef]
- Johannsen, O.A. Revision of the North American Species of the Genus Pentaneura [Tendipedidae: Chironomidae, Diptera]. J. N. Y. Entomol. Soc. 1946, 54, 267–289. [Google Scholar]
- Freeman, P. A study of the Chironomidae (Diptera) of Africa south of the Sahara. Part I. Bull. Brit. Mus. Nat. Hist. Ent. 1955, 4, 1–67. [Google Scholar]
- Roback, S.S. The Subgenus Ablabesmyia of Pentaneura (Diptera; Tendipedidae; Pelopiinae). Trans. Am. Entomol. Soc. 1959, 85, 113–135. [Google Scholar]
- Fittkau, E.J. Die Tanypodinae (Diptera: Chironomidae) (Die Tribus Anatopyggiini, Macropelopiini und Pentaneurini). Abhandlungen zur Larvalsystematik der Insekten 1962, 6, 1–453. [Google Scholar]
- Aburaya, F.H.; Callil, C.T. Variação temporal de larvas de Chironomidae (Diptera) no Alto Rio Paraguai Cáceres, Mato Grosso, Brasil. Revista Brasileira de Zoologia 2007, 24, 565–572. [Google Scholar] [CrossRef]
- Silva, F.L.D.; Ruiz, S.S.; Bochini, G.L.; Moreira, D.C. Functional feeding habits of Chironomidae larvae (Insecta, Diptera) in a lotic system from Midwestern region of São Paulo State, Brazil. Pan Am. J. Aquat. Sci. 2008, 3, 135–141. [Google Scholar]
- Butakka, C.M.D.M.; Gomes, L.C.; Takeda, A.M. Taxonomic and numeric structure of Chironomidae (Diptera) in different habitats of a Neotropical floodplain. Iheringia Série Zoologia 2014, 104, 314–322. [Google Scholar] [CrossRef] [Green Version]
- Mazão, G.R.; Bispo, P.D.C. The influence of physical instream spatial variability on Chironomidae (Diptera) assemblages in Neotropical streams. Limnologica 2016, 60, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Roback, S.S. The adults of the subfamily Tanypodinae (Pelopiinae) in North America (Diptera: Chironomidae). Monogr. Acad. Nat. Sci. Philad. 1971, 17, 1–410. [Google Scholar]
- Silva, F.L.D.; Ekrem, T. Phylogenetic relationships of nonbiting midges in the subfamily Tanypodinae (Diptera: Chironomidae) inferred from morphology. Syst. Ent. 2016, 41, 73–92. [Google Scholar] [CrossRef]
- Paggi, A.C.; Suarez, D.A. Ablabesmyia reissi, spec. nov., a new species of Tanypodinae from Rio Negro province, Argentina, with descriptions of the adult female and preimaginal stages (Insecta, Diptera, Chironomidae). Spixiana 2000, 23, 259–266. [Google Scholar]
- Oliveira, C.S.N.; Fonseca-Gessner, A.A. New species of Ablabesmyia Johannsen (Diptera, Chironomidae, Tanypodinae) from the Neotropical Region, with description of male adults and immature stages. Revista Brasileira de Zoologia 2006, 23, 740–745. [Google Scholar] [CrossRef]
- Niitsuma, H. Revision of the Japanese Ablabesmyia (Diptera: Chironomidae: Tanypodinae), with descriptions of three new species. Zootaxa 2013, 3664, 479–504. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.S.N.; Fonseca-Gessner, A.A.; Navarro Silvia, M.A. The immature stages of Ablabesmyia (Sartaia) metica Roback, 1983 (Diptera: Chironomidae) with keys to subgenera. Zootaxa 2008, 1808, 61–68. [Google Scholar] [CrossRef]
- Silva, F.L.D.; Dantas, G.P.D.S.; Hamada, N. Description of immature stages of Ablabesmyia cordeiroi Neubern, 2013 (Diptera: Chironomidae: Tanypodinae). Acta Amazonica 2019, 49, 118–121. [Google Scholar] [CrossRef]
- Epler, J.H. Biosystematics of the genus Dicrotendipes Kieffer, 1913 (Diptera: Chironomidae) of the world. Mem. Am. Entomol. Soc. 1988, 36, 1–214. [Google Scholar]
- Sæther, O.A. Glossary of chironomid morphology terminology (Diptera: Chironomidae). Ent. Scand. Supp. 1980, 14, 1–51. [Google Scholar]
- Kowalyk, H.E. The larval cephalic setae in the Tanypodinae (Diptera: Chironomidae) and their importance in generic determinations. Can. Entomol. 1985, 117, 67–106. [Google Scholar] [CrossRef]
- Silva, F.L.D.; Fonseca-Gessner, A.A.; Ekrem, T. Revision of Labrundinia maculata Roback, 1971, a new junior synonym of L. longipalpis (Goetghebuer, 1921) (Diptera: Chironomidae: Tanypodinae). Aquat. Insects 2011, 33, 293–303. [Google Scholar] [CrossRef]
- Da Silva, F.L.; Fonseca-Gessner, A.A.; Ekrem, T. A taxonomic revision of genus Labrundinia Fittkau, 1962 (Diptera: Chironomidae: Tanypodinae). Zootaxa 2014, 3769, 1–185. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Hernández-Triana, L.M.; Prosser, S.W.; Rodríguez-Perez, M.A.; Chaverri, L.G.; Hebert, P.D.N.; Ryan Gregory, T. Recovery of DNA barcodes from blackfly museum specimens (Diptera: Simuliidae) using primer sets that target a variety of sequence lengths. Mol. Ecol. Resour. 2014, 14, 508–518. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the boostrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Sublette, J.E.; Sublette, M.S. Family Chironomidae (Tendipedidae). In A Catalog of the Diptera of America North of Mexico; Stone, A., Sabrosky, C.W., Wirth, W.W., Foote, R.H., Coulson, J.R., Eds.; United States Department of Agriculture: Washington, DC, USA, 1965. [Google Scholar]
- Linnaeus, C. Systema Naturæ per Regna Tria Naturæ, Secundum Classses, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis; Laurentius Salvius: Stockholm, Sweden, 1758. [Google Scholar]
- Prie, V.; Puillandre, N.; Bouchet, P. Bad taxonomy can kill: Molecular reevaluation of Unio mancus Lamarck, 1819 (Bivalvia: Unionidae) and its accepted subspecies. Knowl. Manag. Aquat. Ec. 2012, 08. [Google Scholar] [CrossRef]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Pereira, L.H.G.; Pazian, M.F.; Hanner, R.; Foresti, F.; Oliveira, C. DNA barcoding reveals hidden diversity in the Neotropical freshwater fish Piabina argentea (Characiformes: Characidae) from the Upper Paraná Basin of Brazil. Mitochondr. DNA 2011, 22, 87–96. [Google Scholar] [CrossRef]
- Ekrem, T.; Stur, E.; Orton, M.G.; Adamowicz, S.J. DNA barcode data reveal biogeographic trends in Arctic non-biting midges. Genome 2018, 61, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Lin, X.L.; Wang, Q.; Wang, X.H. DNA barcodes successfully delimit morphospecies in a superdiverse insect genus. Zool. SCR 2018, 47, 311–324. [Google Scholar] [CrossRef]
- Tokunaga, M. Chironomidae from Japan (Diptera), IX. Tanypodinae and Diamesinae. Philipp. J. Sci. 1937, 62, 21–63. [Google Scholar]
- Kobayashi, T.; Kubota, K. A revision of male adult Ablabesmyia (Diptera: Chironomidae: Tanypodinae) from Japan, with a description of A. prorasha, new species, and a key to adult male species of the genus. Raffles Bull. Zool. 2002, 50, 317–326. [Google Scholar]
- Chaudhuri, P.K.; Debnath, R.K.; Nandi, S.K. Tanypodine midges of the genus Ablabesmyia Johannsen (Diptera: Chironomidae) from West Bengal with a note on their seasonal incidence and sex ratios. J. Nat. Hist. 1983, 17, 901–917. [Google Scholar] [CrossRef]
- Sæther, O.A.; Spies, M. Fauna Europaea: Chironomidae. In Fauna Europaea: Diptera: Nematocera; De Jong, H., Pape, T., Eds.; Fauna Europaea: Berlin, Germany, 2013; Available online: https://fauna-eu.org (accessed on 18 September 2019).
- Bernet, G.P.; Bramardi, S.; Calvache, D.; Carbonell, E.A.; Asins, M.J. Applicability of molecular markers in the context of protection of new varieties of cucumber. Plant Breed. 2003, 122, 146–152. [Google Scholar] [CrossRef]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
- Kress, W.J.; García-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2015, 30, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Carew, M.E.; Pettigrove, V.; Cox, R.L.; Hoffmann, A.A. DNA identification of urban Tanytarsini chironomids (Diptera: Chironomidae). J. N. Am. Benth. Soc. 2007, 26, 587–600. [Google Scholar] [CrossRef]
- Da Silva, F.L.; Wiedenbrug, S.; Trivinho-Strixino, S.; da Oliveira, C.S.N.; Pepinelli, M. Two new species of Hudsonimyia Roback, 1979 (Diptera: Chironomidae: Tanypodinae) from Neotropical Region unveiled by morphology and DNA barcoding. J. Nat. Hist. 2012, 46, 1615–1638. [Google Scholar] [CrossRef]
- Stur, E.; Ekrem, T. A review of Norwegian Gymnometriocnemus (Diptera, Chironomidae) including the description of two new species and a new name for Gymnometriocnemus volitans (Goetghebuer) sensu Brundin. ZooKeys 2015, 508, 127–142. [Google Scholar] [CrossRef]
- Song, C.; Wang, Q.; Zhang, R.; Sun, B.; Wang, X. Exploring the utility of DNA barcoding in species delimitation of Polypedilum (Tripodura) non-biting midges (Diptera: Chironomidae). Zootaxa 2016, 4079, 534–550. [Google Scholar] [CrossRef]




| Taxon | Country | Life Stage | BOLD ID | NTNU-VM | GB Accession |
|---|---|---|---|---|---|
| Ablabesmyia americana | Canada | Adult | 10PROBE-11666 | JF875917 | |
| Ablabesmyia americana | Canada | Adult | 10PROBE-12218 | JF876334 | |
| Ablabesmyia americana | Canada | Adult | 10PROBE-12910 | JF876913 | |
| Ablabesmyia americana | Canada | Adult | 10PROBE-16464 | KR655494 | |
| Ablabesmyia americana | Canada | Male adult | CHIR_CH10 | 201777 | MK403766 |
| Ablabesmyia americana | Canada | Male adult | CHIR_CH174 | 201778 | MK403744 |
| Ablabesmyia americana | Canada | Male adult | CHIR_CH228 | 201779 | MK403740 |
| Ablabesmyia americana | Canada | Male adult | CHIR_CH267 | 201780 | MK403767 |
| Ablabesmyia americana | Canada | Female adult | CHIR_CH268 | 201781 | MK403747 |
| Ablabesmyia americana | Canada | Male adult | CHIR_CH396 | 201782 | MK403752 |
| Ablabesmyia americana | Canada | Male adult | HLC-27231 | 201785 | KR433631 |
| Ablabesmyia americana | Canada | Female adult | HLC-27246 | 201786 | KR433665 |
| Ablabesmyia americana | Canada | Male adult | HLC-27252 | 201790 | KR443910 |
| Ablabesmyia aspera | Canada | Adult | 10PROBE-10650 | JF875206 | |
| Ablabesmyia aspera | Canada | Adult | 10PROBE-11271 | JF875757 | |
| Ablabesmyia aspera | Canada | Adult | 10PROBE-12725 | JF876793 | |
| Ablabesmyia aspera | Canada | Adult | 10PROBE-12912 | JF876915 | |
| Ablabesmyia aspera | Canada | Adult | 10PROBE-13831 | KR668901 | |
| Ablabesmyia aspera | Norway | Male adult | ATNA539 | 201788 | MK403756 |
| Ablabesmyia aspera | Norway | Male adult | CH-OSF172 | 143691 | JN285991 |
| Ablabesmyia aspera | Norway | Male adult | CH-OSF174 | 143693 | MK403755 |
| Ablabesmyia aspera | Canada | Male adult | CHIR_CH638 | 201783 | MK403753 |
| Ablabesmyia aspera | Canada | Male adult | HLC-27096 | 201784 | KR435725 |
| Ablabesmyia aspera | Norway | Female adult | SOE94 | 124714 | HQ105015 |
| Ablabesmyia longistyla | Norway | Male adult | TRD-CH185 | 200177 | MK403739 |
| Ablabesmyia monilis | Norway | Female adult | ATNA330 | 201789 | HM421433 |
| Ablabesmyia monilis | Norway | Female adult | ATNA538 | 201787 | MK403749 |
| Ablabesmyia monilis | Japan | Female adult | ES358 | 201791 | MK403757 |
| Ablabesmyia monilis | Japan | Male adult | ES360 | 201793 | MK403737 |
| Ablabesmyia monilis | Japan | Male adult | ES361 | 201794 | MK403738 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark213 | 136311 | JF870860 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark214 | 136312 | JF870861 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark258 | 136355 | JN285993 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark458 | 136554 | JN286007 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark508 | 136603 | JN286012 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark568 | 136663 | JF870930 |
| Ablabesmyia monilis | Norway | Male adult | Finnmark721 | 136814 | JN286021 |
| Ablabesmyia monilis | Norway | Male adult | NO 104 | 200861 | MK403758 |
| Ablabesmyia monilis | Norway | Female adult | NO 92 | 200850 | MK403743 |
| Ablabesmyia monilis | Norway | Male adult | RIN_CH13 | 148138 | MK403765 |
| Ablabesmyia monilis | Norway | Male adult | RIN_CH15 | 148140 | MK403760 |
| Ablabesmyia monilis | Norway | Male adult | RIN_CH29 | 148153 | MK403748 |
| Ablabesmyia monilis | Norway | Larva | RIN_CH30 | 148154 | MK403741 |
| Ablabesmyia monilis | Norway | Larva | RIN_CH32 | 148156 | MK403754 |
| Ablabesmyia monilis | Norway | Larva | RIN_CH34 | 148158 | MK403764 |
| Ablabesmyia monilis | Norway | Larva | RIN_CH50 | 148174 | MK403759 |
| Ablabesmyia monilis | Norway | Male adult | SOE10 | 124308 | HQ105016 |
| Ablabesmyia monilis | Norway | Male adult | SOE9 | 124307 | HQ105017 |
| Ablabesmyia monilis | Norway | Male adult | TRD-CH158 | 200150 | MK403761 |
| Ablabesmyia monilis | Norway | Female adult | TRD-CH221 | 200211 | MK403750 |
| Ablabesmyia monilis | Norway | Male adult | TRD-CH292 | 200278 | MK403763 |
| Ablabesmyia monilis | Norway | Male adult | TRD-CH308 | 200294 | MK403762 |
| Ablabesmyia monilis | Norway | Female adult | TRD-CH309 | 200295 | MK403751 |
| Ablabesmyia monilis | Norway | Male adult | TRD-CH325 | 200310 | MK403742 |
| Ablabesmyia monilis | Norway | Male pupa | TRD-CH427 | 200410 | MK403745 |
| Ablabesmyia monilis | Norway | Female adult | TRD-CH8 | 148261 | MK403746 |
| Nucleotide Position | Variable Sites (%) | Informative Sites (%) | T (%) | C (%) | A (%) | G (%) | AT (%) | GC (%) |
|---|---|---|---|---|---|---|---|---|
| 1st | 15.2 | 13.9 | 27.2 | 16 | 26.4 | 30.4 | 53.6 | 46.4 |
| 2nd | 1.9 | 0.0 | 43 | 27.4 | 13.6 | 16 | 56.6 | 43.4 |
| 3rd | 82.9 | 86.1 | 47 | 8.4 | 42.2 | 2.4 | 89.2 | 10.8 |
| All | 24.9% | 83.5% | 39.1 | 17.3 | 27.4 | 16.2 | 66.5 | 33.5 |
| Species | Average Intraspecific K2-P Divergence | Maximum Intraspecific K2-P Divergence | K2-P Divergence to Nearest Neighbor | Average Interspecific K2-P Divergence |
|---|---|---|---|---|
| Ablabesmyia aspera | 3.7% | 8.0% | 10.8% | 12.6% |
| Ablabesmyia americana | 0.6% | 1.3% | 10.0% | 11.9% |
| Ablabesmyia longistyla | N/A | N/A | 14.4% | 16.2% |
| Ablabesmyia monilis | 4.2% | 8.6% | 10.0% | 12.5% |
| Leg Position | fe | ti | ta1 | ta2 | ta3 |
| p1 | 837 | 924 | 743 | 450 | 329 |
| p2 | 985–1126 | 982–1125 | 778 | 508 | 329 |
| p3 | 898–1075 | 1226–1381 | 1137 | 657 | 473 |
| Leg Position | ta4 | ta5 | LR | BV | SV |
| p1 | 229 | 153 | 0.80 | 2.15 | 2.37 |
| p2 | 227 | 168 | 0.69 | 2.46 | 2.89 |
| p3 | 291 | 181 | 0.82 | 2.24 | 2.16 |
| Leg Position | Fe | ti | ta1 | ta2 | ta3 |
| p1 | 1134 | 1383 | 1079 | 661 | 480 |
| p2 | 1114–1397 | 887–1413 | 556–1028 | 557–626 | 386–456 |
| p3 | 1163–1174 | 1177–1246 | 876–1492 | 500–908 | 341–637 |
| Leg Position | ta4 | ta5 | LR | BV | SV |
| p1 | 307 | 210 | 0.78 | 2.17 | 2.33 |
| p2 | 261–297 | 177–196 | 0.63–0.73 | 1.85–2.44 | 2.73–3.60 |
| p3 | 230-380 | 153–224 | 0.74–1.20 | 1.82–2.63 | 1.62–2.67 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stur, E.; da Silva, F.L.; Ekrem, T. Back from the Past: DNA Barcodes and Morphology Support Ablabesmyia americana Fittkau as a Valid Species (Diptera: Chironomidae). Diversity 2019, 11, 173. https://doi.org/10.3390/d11090173
Stur E, da Silva FL, Ekrem T. Back from the Past: DNA Barcodes and Morphology Support Ablabesmyia americana Fittkau as a Valid Species (Diptera: Chironomidae). Diversity. 2019; 11(9):173. https://doi.org/10.3390/d11090173
Chicago/Turabian StyleStur, Elisabeth, Fabio Laurindo da Silva, and Torbjørn Ekrem. 2019. "Back from the Past: DNA Barcodes and Morphology Support Ablabesmyia americana Fittkau as a Valid Species (Diptera: Chironomidae)" Diversity 11, no. 9: 173. https://doi.org/10.3390/d11090173