cpDNA Barcoding by Combined End-Point and Real-Time PCR Analyses to Identify and Quantify the Main Contaminants of Oregano (Origanum vulgare L.) in Commercial Batches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Nucleic Acid Purification
2.3. Barcode Regions Amplification, Cloning and Sequencing
2.4. Quantitative PCR (qPCR)
3. Results and Discussion
3.1. Qualitative Determination of Contaminants in Oregano Samples
3.2. Quantitative Determination of Contaminants on Oregano Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; de Mattia, F. DNA Barcoding for Minor Crops and Food Traceability. Adv. Agric. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Woolfe, M.; Primrose, S. Food forensics: Using DNA technology to combat misdescription and fraud. Trends Biotechnol. 2004, 22, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Dhanya, K.; Sasikumar, B. Molecular marker based adulteration detection in traded food and agricultural commodities of plant origin with special reference to spices. Curr. Trends Biotechnol. Pharm. 2010, 4, 454–489. [Google Scholar]
- Posadzki, P.; Watson, L.; Ernst, E. Contamination and adulteration of herbal medicinal products (HMPs): An overview of systematic reviews. Eur. J. Clin. Pharmacol. 2013, 69, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Galimberti, A.; Bartolucci, F.; Bruni, I.; de mattia, F.; Cortis, P.; Labra, M. DNA barcoding to analyse taxonomically complex groups in plants: The case of Thymus (Lamiaceae). Bot. J. Linn. Soc. 2013, 171, 687–699. [Google Scholar] [CrossRef]
- Gismondi, A.; Fanali, F.; Martínez Labarga, J.M.; Caiola, M.G.; Canini, A. Crocus sativus L. Genomics and different DNA barcode applications. Plant Syst. Evol. 2013, 299, 1859–1863. [Google Scholar] [CrossRef]
- Kojoma, M.; Kurihara, K.; Yamada, K.; Sekita, S.; Satake, M.; Iida, O. Genetic identification of cinnamon (Cinnamomum spp.) based on the trnL-trnF chloroplast DNA. Planta Med. 2002, 68, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, H.X.; Wang, L.; Wang, T.; Yang, R.W.; Wang, X.L.; Zhou, Y.H.; Ding, C.B.; Zhang, L. Potential use of DNA barcoding for the identification of Salvia based on cpDNA and nrDNA sequences. Gene 2013, 528, 206–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marieschi, M.; Torelli, A.; Poli, F.; Sacchetti, G.; Bruni, R. RAPD-Based method for the quality control of mediterranean oregano and its contribution to pharmacognostic techniques. J. Agric. Food Chem. 2009, 57, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Kintzios, S.E. Medicinal and aromatic plants—Industrial profiles. In Oregano: The Genera Origanum and Lippia; CRC Press: London, UK, 2002; Volume 25, pp. 3–9. ISBN 0-415-36943-6. [Google Scholar]
- Skoula, M.; Harborne, J.B. The taxonomy and chemistry of Origanum. In Oregano: The Genera Origanum and Lippia; Kintzios, S.E., Ed.; Taylor & Francis, CRC Press: London, UK, 2002; pp. 67–108. [Google Scholar]
- Ietswaart, J.H. A Taxonomic Revision of the Genus Origanum (Labiatae); Leiden University Press: Leiden, The Netherlands, 1980; Volume 4. [Google Scholar]
- Baricevic, D.; Bartol, T. The biological/pharmacological activity of the Origanum genus. In Oregano: The Genera Origanum and Lippia; Kintzios, S.E., Ed.; Taylor & Francis: London, UK, 2002; Chapter 8; pp. 177–213. [Google Scholar]
- Tucker, A.O.; DeBaggio, T. The Big Book of Herbs: A Comprehensive Illustrated Reference to Herbs of Flavor and Fragrance; Interweave Press: Loveland, CO, USA, 2000. [Google Scholar]
- Campanini, E. Dizionario di Fitoterapia e Piante Medicinali; Tecniche Nuove: Milano, Italy, 1998. [Google Scholar]
- Kintzios, S.E. Oregano. In Handbook of Herbs and Spices, 1st ed.; Peter, K.V., Ed.; Woodhead Publishing: Cambridge, UK, 2004; Volume 2, pp. 215–229. [Google Scholar]
- Tainter, D.R.; Grenis, A.T. The spices. In Spices and Seasonings: A Food Technology Handbook, 2nd ed.; Tainter, D.R., Grenis, A.T., Eds.; Wiley-VCH: New York, NY, USA, 2001; pp. 116–119. [Google Scholar]
- Huie, C.W. A review of modern sample-preparation techniques for the extraction and analysis of medicinal plants. Anal. Bioanal. Chem. 2002, 373, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Schultz, H.; Quilitzsch, R.; Kruger, H. Rapid evaluation and quantitative analysis of thyme, origano and chamomile essential oils by ATR-IR and NIR spectroscopy. J. Mol. Struct. 2003, 661–662, 299–306. [Google Scholar] [CrossRef]
- Techen, N.; Crockett, S.L.; Khan, I.A.; Scheffler, B.E. Authentication of medicinal plants using molecular biology techniques to compliment conventional methods. Curr. Med. Chem. 2004, 11, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA barcoding as a molecular tool to track down mislabeling and food piracy. Diversity 2016, 8. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of Plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, A.; De Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
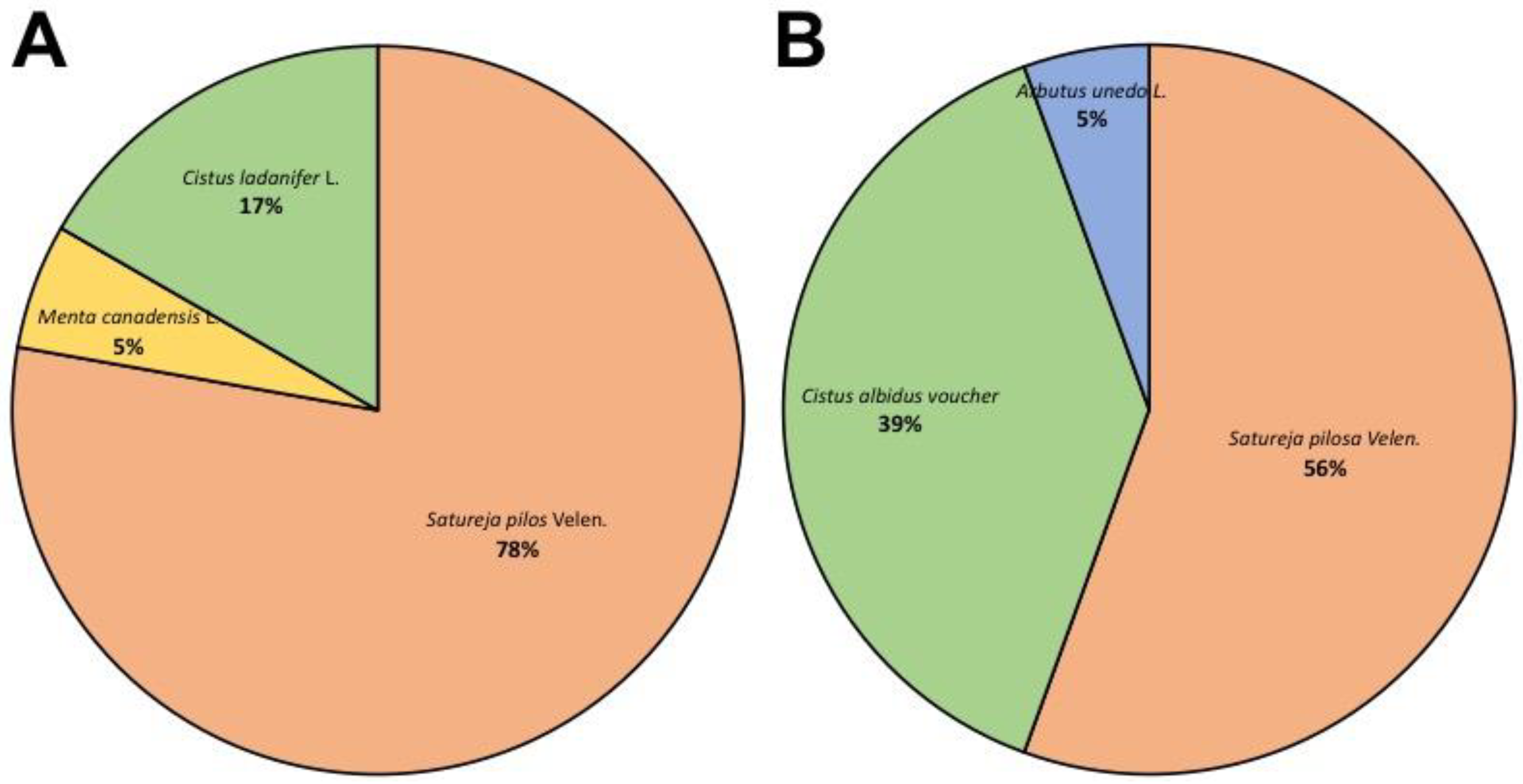
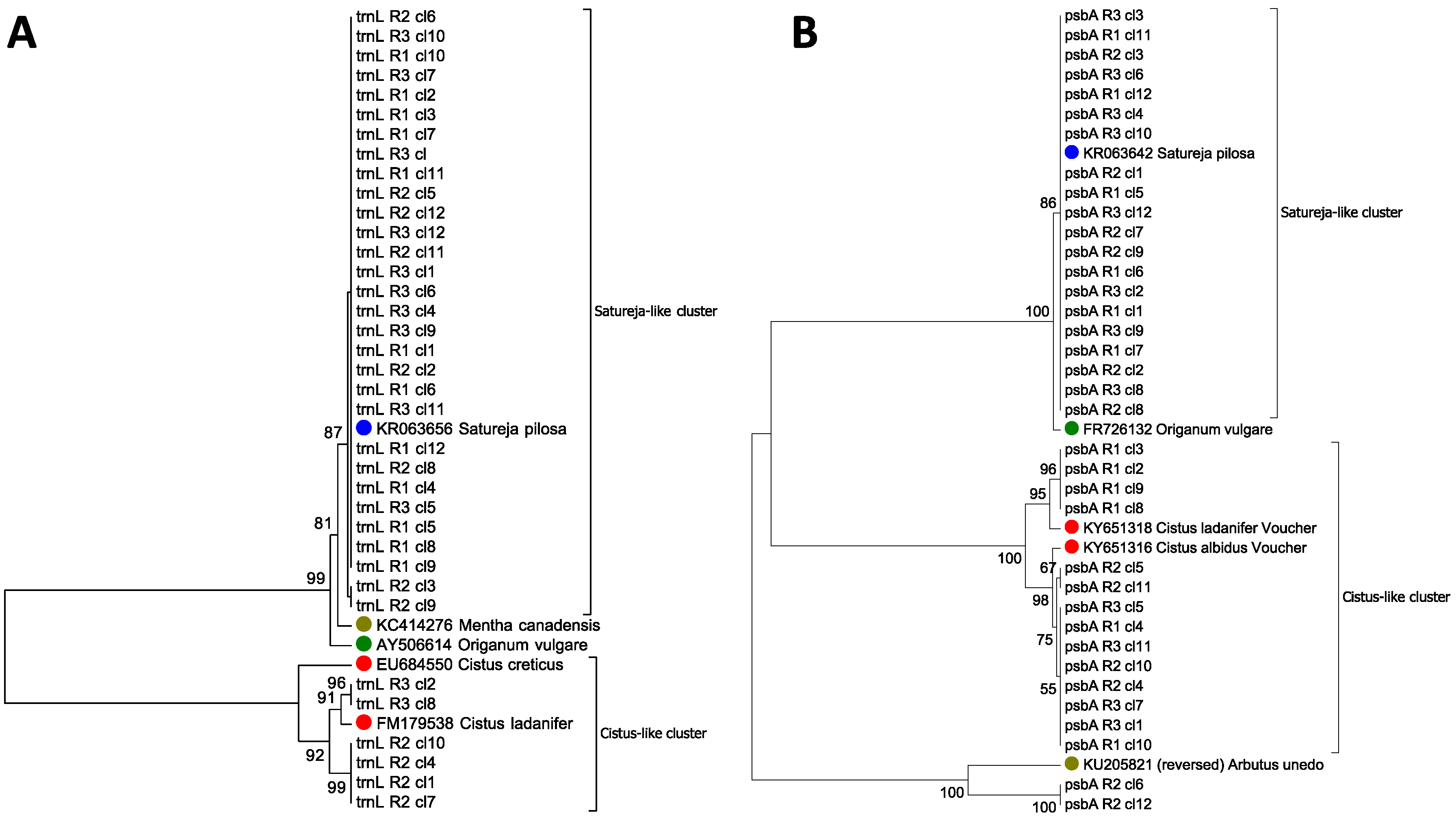
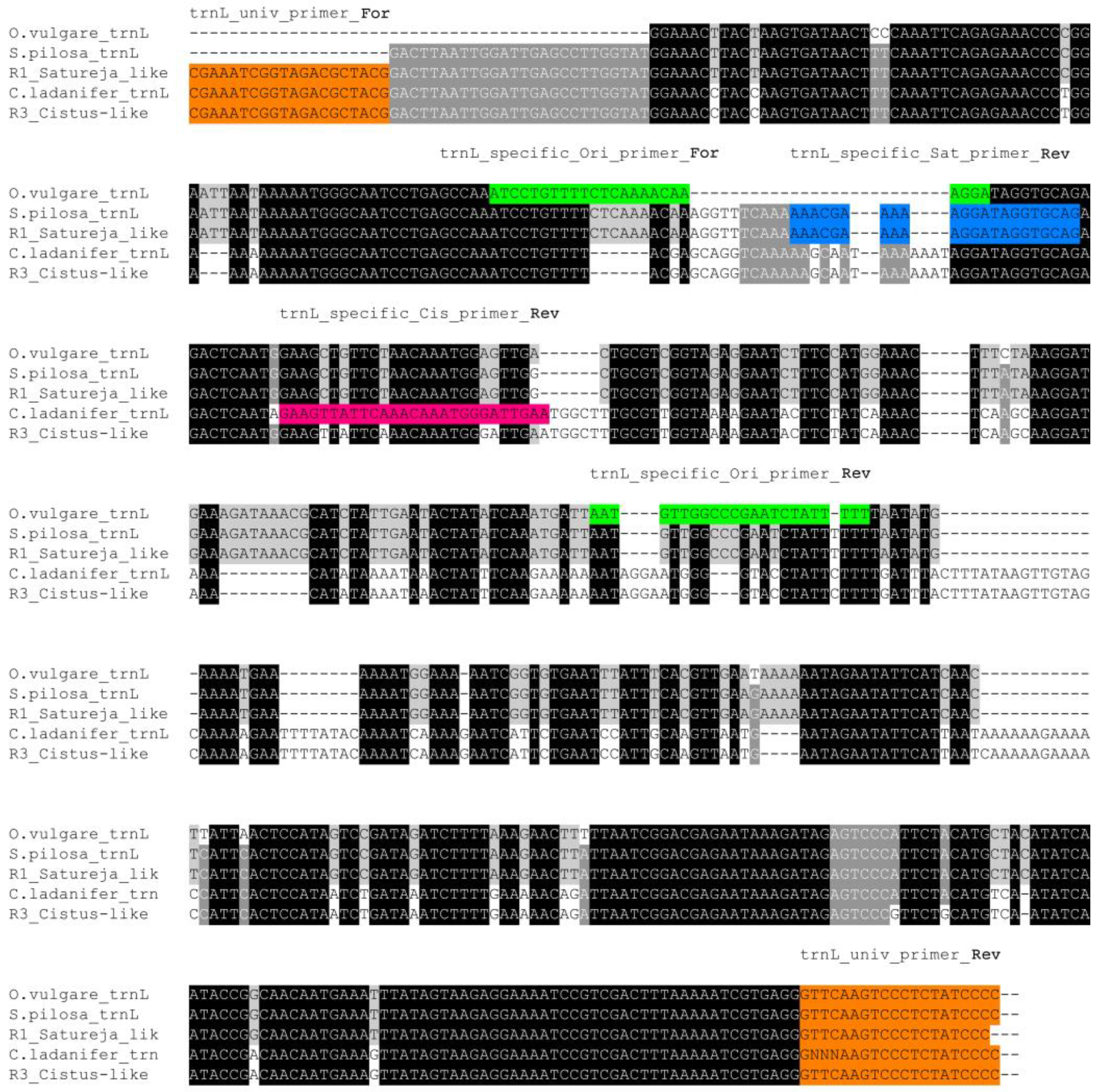
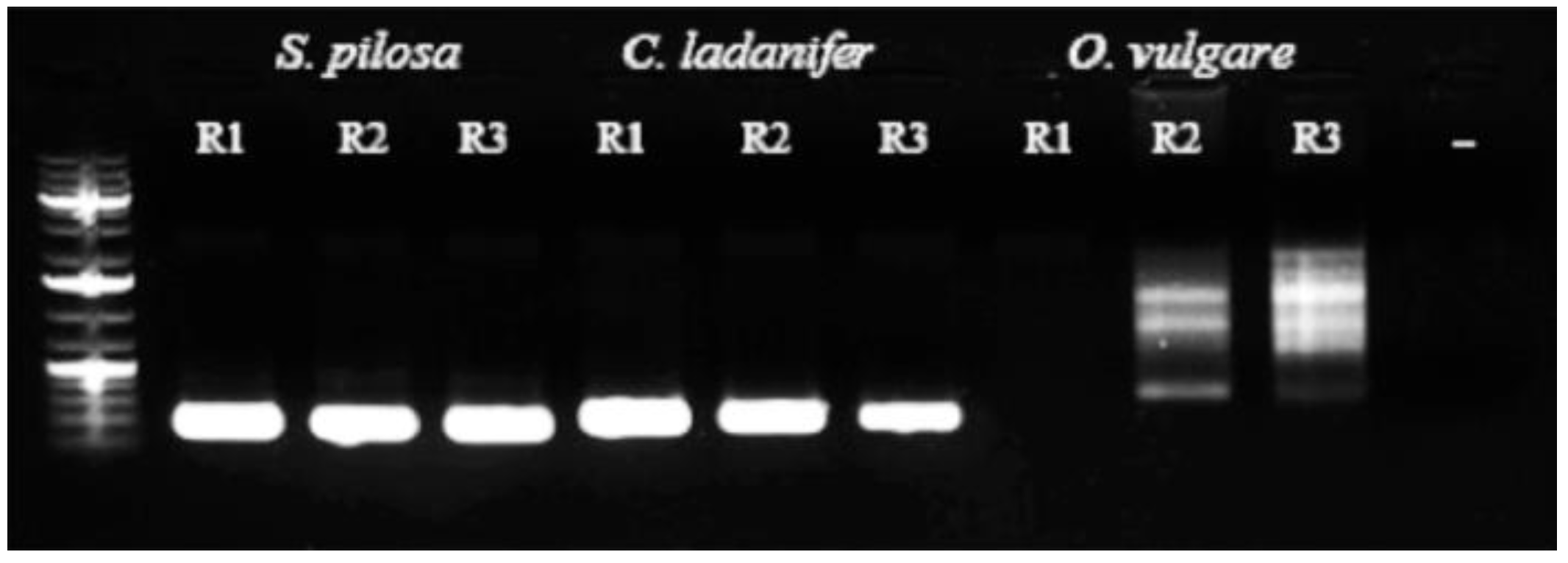
| Name | Best HIT | Organism | Coverage | Identity | E-Value |
|---|---|---|---|---|---|
| trnL_R1_cl1 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl2 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl3 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl4 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.4% | 100% | 0 |
| trnL_R1_cl5 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl6 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl7 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl8 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl9 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl10 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.4% | 100% | 0 |
| trnL_R1_cl11 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R1_cl12 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R2_cl1 | FM179538/EU684550 | Cistus ladanifer L./Cistus creticus L. | 100% | 97.2% | 0 |
| trnL_R2_cl2 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R2_cl3 | AY506611 | Menta canadensis L. | 100% | 98.1% | 0 |
| trnL_R2_cl4 | FM179538/EU684550 | Cistus ladanifer L./Cistus creticus L. | 100% | 97.2% | 0 |
| trnL_R2_cl5 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.4% | 100% | 0 |
| trnL_R2_cl6 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.4% | 100% | 0 |
| trnL_R2_cl7 | FM179538/EU684550 | Cistus ladanifer L./Cistus creticus L. | 100% | 97.2% | 0 |
| trnL_R2_cl8 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R2_cl9 | AY506611 | Menta canadensis L. | 100% | 98.1% | 0 |
| trnL_R2_cl10 | FM179538/EU684550 | Cistus ladanifer L./Cistus creticus L. | 100% | 97.2% | 0 |
| trnL_R2_cl11 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.4% | 100% | 0 |
| trnL_R2_cl12 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.4% | 100% | 0 |
| trnL_R3_cl1 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl2 | FM179538/EU684550 | Cistus ladanifer L./Cistus creticus L. | 100% | 98.2% | 0 |
| trnL_R3_cl3 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl4 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl5 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl6 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl7 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl8 | FM179538/EU684550 | Cistus ladanifer L./Cistus creticus L. | 100% | 98.2% | 0 |
| trnL_R3_cl9 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl10 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl11 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| trnL_R3_cl12 | KR063656/JQ669067 | Satureja pilosa Velen./Satureja montana L. | 96.5% | 100% | 0 |
| Name | Best HIT | Organism | Coverage | Identity | E-Value |
|---|---|---|---|---|---|
| trnH-psbA_R1_cl1 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R1_cl2 | KY651318 | Cistus ladanifer voucher SEV:286741 | 46.3% | 98.3% | 2.62e-108 |
| trnH-psbA_R1_cl3 | KY651318 | Cistus ladanifer voucher SEV:286741 | 46.95% | 98.3% | 2.058e-108 |
| trnH-psbA_R1_cl4 | KY651316 | Cistus albidus voucher SEV:286739 | 55.98% | 99.1% | 2.15e-113 |
| trnH-psbA_R1_cl5 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R1_cl6 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R1_cl7 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R1_cl8 | KY651318 | Cistus ladanifer voucher SEV:286741 | 46.3% | 98.3% | 2.62e-108 |
| trnH-psbA_R1_cl9 | KY651318 | Cistus ladanifer voucher SEV:286741 | 46.95% | 98.3% | 2.058e-108 |
| trnH-psbA_R1_cl10 | KY651316 | Cistus albidus voucher SEV:286739 | 55.98% | 99.1% | 2.15e-113 |
| trnH-psbA_R1_cl11 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R1_cl12 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R2_cl1 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R2_cl2 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R2_cl3 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.0% | 100% | 0 |
| trnH-psbA_R2_cl4 | KY651316 | Cistus albidus voucher SEV:286739 | 55.95% | 98.7% | 2.79e-112 |
| trnH-psbA_R2_cl5 | KY651316 | Cistus albidus voucher SEV:286739 | 55.9% | 99.1% | 7.67e-113 |
| trnH-psbA_R2_cl6 | KU205821 | Arbutus uneto L. | 73.4% | 100% | 1.92e-162 |
| trnH-psbA_R2_cl7 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R2_cl8 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R2_cl9 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.0% | 100% | 0 |
| trnH-psbA_R2_cl10 | KY651316 | Cistus albidus voucher SEV:286739 | 55.95% | 98.7% | 2.79e-112 |
| trnH-psbA_R2_cl11 | KY651316 | Cistus albidus voucher SEV:286739 | 55.9% | 99.1% | 7.67e-113 |
| trnH-psbA_R2_cl12 | KU205821 | Arbutus uneto L. | 73.4% | 100% | 1.92e-162 |
| trnH-psbA_R3_cl1 | KY651316 | Cistus albidus voucher SEV:286739 | 55.98% | 99.1% | 2.15e-113 |
| trnH-psbA_R3_cl2 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 99.7% | 0 |
| trnH-psbA_R3_cl3 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R3_cl4 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.0% | 100% | 0 |
| trnH-psbA_R3_cl5 | KY651316 | Cistus albidus voucher SEV:286739 | 55.74% | 99.6% | 4.62e-115 |
| trnH-psbA_R3_cl6 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R3_cl7 | KY651316 | Cistus albidus voucher SEV:286739 | 55.98% | 99.1% | 2.15e-113 |
| trnH-psbA_R3_cl8 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 99.7% | 0 |
| trnH-psbA_R3_cl9 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| trnH-psbA_R3_cl10 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.0% | 100% | 0 |
| trnH-psbA_R3_cl11 | KY651316 | Cistus albidus voucher SEV:286739 | 55.74% | 99.6% | 4.62e-115 |
| trnH-psbA_R3_cl12 | KR063642 | Satureja pilosa Velen./Satureja montana L. | 88.2% | 100% | 0 |
| Sample Name | Target Name | Ct Mean | Ct SD | Quantity Mean | Quantity SD |
|---|---|---|---|---|---|
| R1 | trnL_Ori | 26.17 | 0.04 | 4.22 | 0.05 |
| R2 | trnL_Ori | 25.37 | 0.05 | 4.01 | 0.13 |
| R3 | trnL_Ori | 25.82 | 0.06 | 3.94 | 0.12 |
| STD 100 | trnL_Ori | 17.53 | 0.08 | ||
| STD 50 | trnL_Ori | 18.60 | 0.14 | ||
| STD 25 | trnL_Ori | 19.74 | 0.05 | ||
| STD 12.5 | trnL_Ori | 20.80 | 0.11 | ||
| STD 6.25 | trnL_Ori | 21.73 | 0.05 | ||
| STD 3.125 | trnL_Ori | 32.97 | 0.26 | ||
| R1 | trnL_Cis | 16.33 | 0.03 | 6.03 | 0.05 |
| R2 | trnL_Cis | 17.53 | 0.09 | 5.48 | 0.16 |
| R3 | trnL_Cis | 15.80 | 0.49 | 6.23 | 0.64 |
| STD 100 | trnL_Cis | 10.03 | 0.09 | ||
| STD 50 | trnL_Cis | 10.84 | 0.06 | ||
| STD 25 | trnL_Cis | 11.91 | 0.03 | ||
| STD 12.5 | trnL_Cis | 13.23 | 0.09 | ||
| STD 6.25 | trnL_Cis | 14.12 | 0.11 | ||
| STD 3.125 | trnL_Cis | 30.44 | 0.12 | ||
| R1 | trnL_Sat | 17.79 | 0.17 | 35.97 | 1.91 |
| R2 | trnL_Sat | 17.88 | 0.05 | 34.93 | 0.57 |
| R3 | trnL_Sat | 18.53 | 2.30 | 33.43 | 10.07 |
| STD 100 | trnL_Sat | 15.97 | 0.24 | ||
| STD 50 | trnL_Sat | 17.03 | 0.16 | ||
| STD 25 | trnL_Sat | 18.15 | 0.38 | ||
| STD 12.5 | trnL_Sat | 19.52 | 0.11 | ||
| STD 6.25 | trnL_Sat | 20.44 | 0.11 | ||
| STD 3.125 | trnL_Sat | 29.64 | 1.12 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vannozzi, A.; Lucchin, M.; Barcaccia, G. cpDNA Barcoding by Combined End-Point and Real-Time PCR Analyses to Identify and Quantify the Main Contaminants of Oregano (Origanum vulgare L.) in Commercial Batches. Diversity 2018, 10, 98. https://doi.org/10.3390/d10030098
Vannozzi A, Lucchin M, Barcaccia G. cpDNA Barcoding by Combined End-Point and Real-Time PCR Analyses to Identify and Quantify the Main Contaminants of Oregano (Origanum vulgare L.) in Commercial Batches. Diversity. 2018; 10(3):98. https://doi.org/10.3390/d10030098
Chicago/Turabian StyleVannozzi, Alessandro, Margherita Lucchin, and Gianni Barcaccia. 2018. "cpDNA Barcoding by Combined End-Point and Real-Time PCR Analyses to Identify and Quantify the Main Contaminants of Oregano (Origanum vulgare L.) in Commercial Batches" Diversity 10, no. 3: 98. https://doi.org/10.3390/d10030098






