Emerging Roles of Vitamin B12 in Aging and Inflammation
Abstract
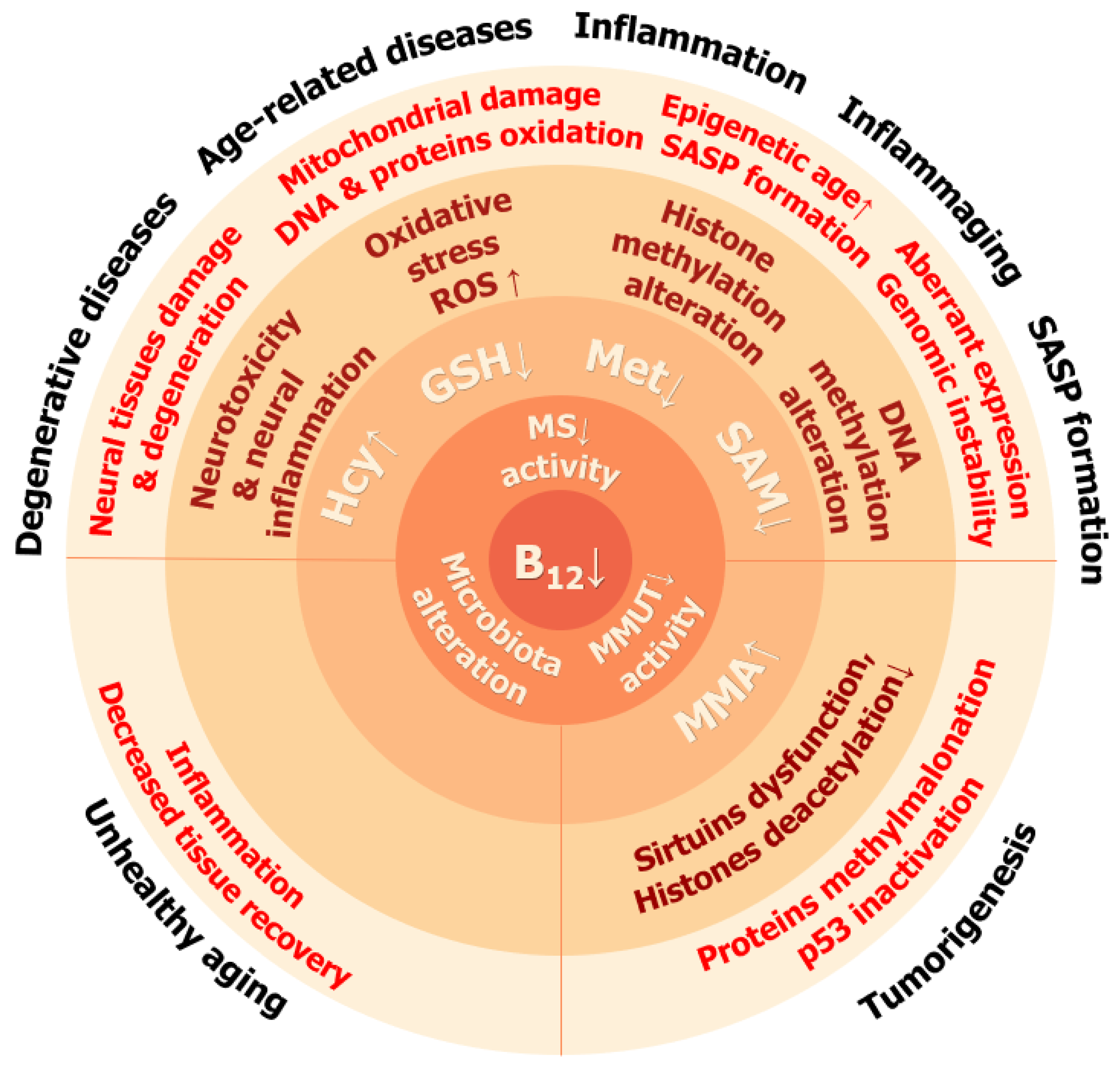
:1. Introduction
2. Vitamin B12 Transport
3. B12-Dependent Proteins
3.1. Methylmalonyl-CoA Mutase
3.2. Methionine Synthase
4. Nucleic Acids and Histone Methylation
5. Consequences of Vitamin B12 Deficiency
6. Role of B12 in Aging, Senescence, and Inflammation
6.1. Vitamin B12 and Its Role in the Formation of the Senescence Phenotype
6.2. Vitamin B12 and Epigenetic Age
6.3. Role of B12 in Cytokine Regulation and Inflammation
6.4. Microbiota, Vitamin B12, and Aging
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kok, D.E.G.; Dhonukshe-Rutten, R.A.M.; Lute, C.; Heil, S.G.; Uitterlinden, A.G.; van der Velde, N.; van Meurs, J.B.J.; van Schoor, N.M.; Hooiveld, G.J.E.J.; de Groot, L.C.P.G.M.; et al. The Effects of Long-Term Daily Folic Acid and Vitamin B12 Supplementation on Genome-Wide DNA Methylation in Elderly Subjects. Clin. Epigenet. 2015, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Head, P.E.; Myung, S.; Chen, Y.; Schneller, J.L.; Wang, C.; Duncan, N.; Hoffman, P.; Chang, D.; Gebremariam, A.; Gucek, M.; et al. Aberrant Methylmalonylation Underlies Methylmalonic Acidemia and Is Attenuated by an Engineered Sirtuin. Sci. Transl. Med. 2022, 14, eabn4772. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Taga, M.E.; Goodman, A.L. Vitamin B12 as a Modulator of Gut Microbial Ecology. Cell Metab. 2014, 20, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, V.; Chabrun, F.; Lacout, C.; Ghali, A.; Capitain, O.; Patsouris, A.; Lavigne, C.; Urbanski, G. Persistent Elevation of Plasma Vitamin B12 Is Strongly Associated with Solid Cancer. Sci. Rep. 2021, 11, 13361. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-López, I.; Kovatcheva, M.; Casas, R.; Toledo, E.; Fitó, M.; Ros, E.; Estruch, R.; Serrano, M.; Lamuela-Raventós, R.M. Higher Circulating Vitamin B12 Is Associated with Lower Levels of Inflammatory Markers in Individuals at High Cardiovascular Risk and in Naturally Aged Mice. J. Sci. Food Agric. 2024, 104, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.F.; Ward, M.; Hoey, L.; McNulty, H. Vitamin B12 and Ageing: Current Issues and Interaction with Folate. Ann. Clin. Biochem. 2013, 50, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, K.; Apostolopoulos, V. B Vitamins and Ageing. In Biochemistry and Cell Biology of Ageing: Part I Biomedical Science; Harris, J.R., Korolchuk, V.I., Eds.; Subcellular Biochemistry; Springer: Singapore, 2018; Volume 90, pp. 451–470. ISBN 9789811328343. [Google Scholar]
- Woodward, R.B. The Total Synthesis of Vitamin B12. Pure Appl. Chem. 1973, 33, 145–178. [Google Scholar] [CrossRef]
- Martens, H.; Barg, M.; Warren, D.; Jah, J.-H. Microbial Production of Vitamin B12. Appl. Microbiol. Biotechnol. 2002, 58, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Barry, N.A.; Mok, K.C.; Taga, M.E.; Goodman, A.L. Human Gut Microbes Use Multiple Transporters to Distinguish Vitamin B12 Analogs and Compete in the Gut. Cell Host Microbe 2014, 15, 47–57. [Google Scholar] [CrossRef]
- Kirmiz, N.; Galindo, K.; Cross, K.L.; Luna, E.; Rhoades, N.; Podar, M.; Flores, G.E. Comparative Genomics Guides Elucidation of Vitamin B12 Biosynthesis in Novel Human-Associated Akkermansia Strains. Appl. Environ. Microbiol. 2020, 86, e02117-19. [Google Scholar] [CrossRef]
- Balabanova, L.; Averianova, L.; Marchenok, M.; Son, O.; Tekutyeva, L. Microbial and Genetic Resources for Cobalamin (Vitamin B12) Biosynthesis: From Ecosystems to Industrial Biotechnology. Int. J. Mol. Sci. 2021, 22, 4522. [Google Scholar] [CrossRef] [PubMed]
- Fedosov, S.N.; Fedosova, N.U.; Kräutler, B.; Nexø, E.; Petersen, T.E. Mechanisms of Discrimination between Cobalamins and Their Natural Analogues during Their Binding to the Specific B12-Transporting Proteins. Biochemistry 2007, 46, 6446–6458. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.J.; Rasmussen, M.R.; Andersen, C.B.F.; Nexø, E.; Moestrup, S.K. Vitamin B12 Transport from Food to the Body’s Cells—A Sophisticated, Multistep Pathway. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 345–354. [Google Scholar] [CrossRef]
- Quadros, E.V.; Nakayama, Y.; Sequeira, J.M. The Protein and the Gene Encoding the Receptor for the Cellular Uptake of Transcobalamin-Bound Cobalamin. Blood 2009, 113, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of MRNAs Genome-Wide. Mol. Cell 2017, 67, 71–83.e7. [Google Scholar] [CrossRef] [PubMed]
- Birn, H.; Willnow, T.E.; Nielsen, R.; Norden, A.G.W.; Bönsch, C.; Moestrup, S.K.; Nexø, E.; Christensen, E.I. Megalin Is Essential for Renal Proximal Tubule Reabsorption and Accumulation of Transcobalamin-B12. Am. J. Physiol.-Ren. Physiol. 2002, 282, F408–F416. [Google Scholar] [CrossRef] [PubMed]
- Rutsch, F.; Gailus, S.; Miousse, I.R.; Suormala, T.; Sagné, C.; Toliat, M.R.; Nürnberg, G.; Wittkampf, T.; Buers, I.; Sharifi, A.; et al. Identification of a Putative Lysosomal Cobalamin Exporter Altered in the CblF Defect of Vitamin B12 Metabolism. Nat. Genet. 2009, 41, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Coelho, D.; Kim, J.C.; Miousse, I.R.; Fung, S.; du Moulin, M.; Buers, I.; Suormala, T.; Burda, P.; Frapolli, M.; Stucki, M.; et al. Mutations in ABCD4 Cause a New Inborn Error of Vitamin B12 Metabolism. Nat. Genet. 2012, 44, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Gherasim, C.; Banerjee, R. Decyanation of Vitamin B12 by a Trafficking Chaperone. Proc. Natl. Acad. Sci. USA 2008, 105, 14551–14554. [Google Scholar] [CrossRef]
- Hannibal, L.; Kim, J.; Brasch, N.E.; Wang, S.; Rosenblatt, D.S.; Banerjee, R.; Jacobsen, D.W. Processing of Alkylcobalamins in Mammalian Cells: A Role for the MMACHC (CblC) Gene Product. Mol. Genet. Metab. 2009, 97, 260–266. [Google Scholar] [CrossRef]
- Suormala, T.; Baumgartner, M.R.; Coelho, D.; Zavadakova, P.; Kožich, V.; Koch, H.G.; Berghaüser, M.; Wraith, J.E.; Burlina, A.; Sewell, A.; et al. The CblD Defect Causes Either Isolated or Combined Deficiency of Methylcobalamin and Adenosylcobalamin Synthesis. J. Biol. Chem. 2004, 279, 42742–42749. [Google Scholar] [CrossRef]
- Padovani, D.; Labunska, T.; Palfey, B.A.; Ballou, D.P.; Banerjee, R. Adenosyltransferase Tailors and Delivers Coenzyme B12. Nat. Chem. Biol. 2008, 4, 194–196. [Google Scholar] [CrossRef]
- Padovani, D.; Banerjee, R. A G-Protein Editor Gates Coenzyme B12 Loading and Is Corrupted in Methylmalonic Aciduria. Proc. Natl. Acad. Sci. USA 2009, 106, 21567–21572. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Arzamasov, A.A.; Khoroshkin, M.S.; Iablokov, S.N.; Leyn, S.A.; Peterson, S.N.; Novichkov, P.S.; Osterman, A.L. Micronutrient Requirements and Sharing Capabilities of the Human Gut Microbiome. Front. Microbiol. 2019, 10, 1316. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, S.; Ravcheev, D.; de Crécy-Lagard, V.; Thiele, I. Systematic Genome Assessment of B-Vitamin Biosynthesis Suggests Co-Operation among Gut Microbes. Front. Genet. 2015, 6, 129714. [Google Scholar] [CrossRef] [PubMed]
- van Asselt, D.Z.; de Groot, L.C.; van Staveren, W.A.; Blom, H.J.; Wevers, R.A.; Biemond, I.; Hoefnagels, W.H. Role of Cobalamin Intake and Atrophic Gastritis in Mild Cobalamin Deficiency in Older Dutch Subjects. Am. J. Clin. Nutr. 1998, 68, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Pannérec, A.; Migliavacca, E.; De Castro, A.; Michaud, J.; Karaz, S.; Goulet, L.; Rezzi, S.; Ng, T.P.; Bosco, N.; Larbi, A.; et al. Vitamin B12 Deficiency and Impaired Expression of Amnionless during Aging. J. Cachexia Sarcopenia Muscle 2018, 9, 41–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Hodgson, N.W.; Trivedi, M.S.; Abdolmaleky, H.M.; Fournier, M.; Cuenod, M.; Do, K.Q.; Deth, R.C. Decreased Brain Levels of Vitamin B12 in Aging, Autism and Schizophrenia. PLoS ONE 2016, 11, e0146797. [Google Scholar] [CrossRef]
- Cannata, J.J.B.; Focesi, A.; Mazumder, R.; Warner, R.C.; Ochoa, S. Metabolism of Propionic Acid in Animal Tissues. J. Biol. Chem. 1965, 240, 3249–3257. [Google Scholar] [CrossRef]
- Frenkel, E.P.; Kitchens, R.L. Intracellular Localization of Hepatic Propionyl-CoA Carboxylase and Methylmalonyl-CoA Mutase in Humans and Normal and Vitamin B12 Deficient Rats. Br. J. Haematol. 1975, 31, 501–513. [Google Scholar] [CrossRef]
- Wolfle, K.; Michenfelder, M.; Konig, A.; Hull, W.E.; Retey, J. On the Mechanism of Action of Methylmalonyl-CoA Mutase. Change of the Steric Course on Isotope Substitution. Eur. J. Biochem. 1986, 156, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Froese, D.S.; Kochan, G.; Muniz, J.R.C.; Wu, X.; Gileadi, C.; Ugochukwu, E.; Krysztofinska, E.; Gravel, R.A.; Oppermann, U.; Yue, W.W. Structures of the Human GTPase MMAA and Vitamin B12-Dependent Methylmalonyl-CoA Mutase and Insight into Their Complex Formation. J. Biol. Chem. 2010, 285, 38204–38213. [Google Scholar] [CrossRef]
- Marsh, E.N.G.; Meléndez, G.D.R. Adenosylcobalamin Enzymes: Theory and Experiment Begin to Converge. Biochim. Biophys. Acta BBA Proteins Proteom. 2012, 1824, 1154–1164. [Google Scholar] [CrossRef]
- Bassila, C.; Ghemrawi, R.; Flayac, J.; Froese, D.S.; Baumgartner, M.R.; Guéant, J.-L.; Coelho, D. Methionine Synthase and Methionine Synthase Reductase Interact with MMACHC and with MMADHC. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2017, 1863, 103–112. [Google Scholar] [CrossRef]
- The GTEx Consortium; Aguet, F.; Anand, S.; Ardlie, K.G.; Gabriel, S.; Getz, G.A.; Graubert, A.; Hadley, K.; Handsaker, R.E.; Huang, K.H.; et al. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Oltean, S.; Banerjee, R. A B12-Responsive Internal Ribosome Entry Site (IRES) Element in Human Methionine Synthase. J. Biol. Chem. 2005, 280, 32662–32668. [Google Scholar] [CrossRef]
- Chen, L.H.; Liu, M.-L.; Hwang, H.-Y.; Chen, L.-S.; Korenberg, J.; Shane, B. Human Methionine Synthase. J. Biol. Chem. 1997, 272, 3628–3634. [Google Scholar] [CrossRef]
- Mendoza, J.; Purchal, M.; Yamada, K.; Koutmos, M. Structure of Full-Length Cobalamin-Dependent Methionine Synthase and Cofactor Loading Captured in Crystallo. Nat. Commun. 2023, 14, 6365. [Google Scholar] [CrossRef] [PubMed]
- Watkins, M.B.; Wang, H.; Burnim, A.; Ando, N. Conformational Switching and Flexibility in Cobalamin-Dependent Methionine Synthase Studied by Small-Angle X-Ray Scattering and Cryoelectron Microscopy. Proc. Natl. Acad. Sci. USA 2023, 120, e2302531120. [Google Scholar] [CrossRef]
- Bandarian, V.; Pattridge, K.A.; Lennon, B.W.; Huddler, D.P.; Matthews, R.G.; Ludwig, M.L. Domain Alternation Switches B(12)-Dependent Methionine Synthase to the Activation Conformation. Nat. Struct. Biol. 2002, 9, 53–56. [Google Scholar] [CrossRef]
- Jarrett, J.T.; Amaratunga, M.; Drennan, C.L.; Scholten, J.D.; Sands, R.H.; Ludwig, M.L.; Matthews, R.G. Mutations in the B12-Binding Region of Methionine Synthase: How the Protein Controls Methylcobalamin Reactivity. Biochemistry 1996, 35, 2464–2475. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Kozlowski, P.M. Mechanistic Insights for Formation of an Organometallic Co–C Bond in the Methyl Transfer Reaction Catalyzed by Methionine Synthase. J. Phys. Chem. B 2013, 117, 16044–16057. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.E.; Matthews, R.G. Protonation State of Methyltetrahydrofolate in a Binary Complex with Cobalamin-Dependent Methionine Synthase. Biochemistry 2000, 39, 13880–13890. [Google Scholar] [CrossRef] [PubMed]
- Drummond, J.T.; Huang, S.; Blumenthal, R.M.; Matthews, R.G. Assignment of Enzymic Function to Specific Protein Regions of Cobalamin-Dependent Methionine Synthase from Escherichia Coli. Biochemistry 1993, 32, 9290–9295. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.R.; Darnell, A.M.; Reilly, M.F.; Kunchok, T.; Joesch-Cohen, L.; Rosenberg, D.; Ali, A.; Rees, M.G.; Roth, J.A.; Lewis, C.A. Methionine Synthase Is Essential for Cancer Cell Proliferation in Physiological Folate Environments. Nat. Metab. 2021, 3, 1500–1511. [Google Scholar] [CrossRef] [PubMed]
- West, A.A.; Caudill, M.A.; Bailey, L.B. Folate. In Present Knowledge in Nutrition; Elsevier: Amsterdam, The Netherlands, 2020; pp. 239–255. [Google Scholar]
- Hashimoto, H.; Vertino, P.M.; Cheng, X. Molecular Coupling of DNA Methylation and Histone Methylation. Epigenomics 2010, 2, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Tu, B.P. Sink into the Epigenome: Histones as Repositories That Influence Cellular Metabolism. Trends Endocrinol. Metab. 2018, 29, 626–637. [Google Scholar] [CrossRef]
- Ye, C.; Sutter, B.M.; Wang, Y.; Kuang, Z.; Tu, B.P. A Metabolic Function for Phospholipid and Histone Methylation. Mol. Cell 2017, 66, 180–193.e8. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Locasale, J.W.; Lyssiotis, C.A.; Zheng, Y.; Teo, R.Y.; Ratanasirintrawoot, S.; Zhang, J.; Onder, T.; Unternaehrer, J.J.; Zhu, H.; et al. Influence of Threonine Metabolism on S -Adenosylmethionine and Histone Methylation. Science 2013, 339, 222–226. [Google Scholar] [CrossRef]
- Sharma, A.; Kramer, M.L.; Wick, P.F.; Liu, D.; Chari, S.; Shim, S.; Tan, W.; Ouellette, D.; Nagata, M.; DuRand, C.J.; et al. D4 Dopamine Receptor-Mediated Phospholipid Methylation and Its Implications for Mental Illnesses Such as Schizophrenia. Mol. Psychiatry 1999, 4, 235–246. [Google Scholar] [CrossRef]
- Hodgson, N.W.; Waly, M.I.; Trivedi, M.S.; Power-Charnitsky, V.-A.; Deth, R.C. Methylation-Related Metabolic Effects of D4 Dopamine Receptor Expression and Activation. Transl. Psychiatry 2019, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pan, T. Methionine Mistranslation Bypasses the Restraint of the Genetic Code to Generate Mutant Proteins with Distinct Activities. PLoS Genet. 2015, 11, e1005745. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Weiss, S.J.; Levine, R.L. Methionine Oxidation and Reduction in Proteins. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Kuschel, L.; Hansel, A.; Schönherr, R.; Weissbach, H.; Brot, N.; Hoshi, T.; Heinemann, S.H. Molecular Cloning and Functional Expression of a Human Peptide Methionine Sulfoxide Reductase (HMsrA). FEBS Lett. 1999, 456, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Meister, A.; Anderson, M.E. Glutathione. Annu. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.B.; Butterfield, D.A. Measurement of Oxidized/Reduced Glutathione Ratio. In Protein Misfolding and Cellular Stress in Disease and Aging; Bross, P., Gregersen, N., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 648, pp. 269–277. ISBN 978-1-60761-755-6. [Google Scholar]
- Li, E.; Zhang, Y. DNA Methylation in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M.; Wang, R.Y.-H. 5-Methylcytosine in Eukaryotic DNA. Science 1981, 212, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- Menke, A.; Dubini, R.C.A.; Mayer, P.; Rovó, P.; Daumann, L.J. Formation of Cisplatin Adducts with the Epigenetically Relevant Nucleobase 5-Methylcytosine. Eur. J. Inorg. Chem. 2021, 2021, 30–36. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.P.; Jaenisch, R. Non-CpG Methylation Is Prevalent in Embryonic Stem Cells and May Be Mediated by DNA Methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA Methylation on N6-Adenine in Mammalian Embryonic Stem Cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, M.; Guo, M. DNA N6-Methyladenine Epigenetic Modification Elevated in Human Esophageal Squamous Cell Carcinoma: A Potential Prognostic Marker. Discov. Med. 2020, 29, 85–90. [Google Scholar] [PubMed]
- Lin, Q.; Chen, J.; Yin, H.; Li, M.; Zhou, C.; Hao, T.; Pan, T.; Wu, C.; Li, Z.; Zhu, D.; et al. DNA N6-Methyladenine Involvement and Regulation of Hepatocellular Carcinoma Development. Genomics 2022, 114, 110265. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA Methylation of the First Exon Is Tightly Linked to Transcriptional Silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Han, H.; De Carvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Rappolee, D.A.; Ruden, D.M. Epigenetic Reprogramming in Mice and Humans: From Fertilization to Primordial Germ Cell Development. Cells 2023, 12, 1874. [Google Scholar] [CrossRef]
- Bollati, V.; Schwartz, J.; Wright, R.; Litonjua, A.; Tarantini, L.; Suh, H.; Sparrow, D.; Vokonas, P.; Baccarelli, A. Decline in Genomic DNA Methylation through Aging in a Cohort of Elderly Subjects. Mech. Ageing Dev. 2009, 130, 234–239. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Kanazhevskaya, L.Y.; Fedorova, O.S. DNA Demethylation in the Processes of Repair and Epigenetic Regulation Performed by 2-Ketoglutarate-Dependent DNA Dioxygenases. Int. J. Mol. Sci. 2021, 22, 10540. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.-H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.-T.; et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated TRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e16. [Google Scholar] [CrossRef]
- De Cecco, M.; Criscione, S.W.; Peckham, E.J.; Hillenmeyer, S.; Hamm, E.A.; Manivannan, J.; Peterson, A.L.; Kreiling, J.A.; Neretti, N.; Sedivy, J.M. Genomes of Replicatively Senescent Cells Undergo Global Epigenetic Changes Leading to Gene Silencing and Activation of Transposable Elements. Aging Cell 2013, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Jansz, N. DNA Methylation Dynamics at Transposable Elements in Mammals. Essays Biochem. 2019, 63, 677–689. [Google Scholar] [PubMed]
- Yang, Z.; Xu, F.; Wang, H.; Teschendorff, A.E.; Xie, F.; He, Y. Pan-Cancer Characterization of Long Non-Coding RNA and DNA Methylation Mediated Transcriptional Dysregulation. EBioMedicine 2021, 68, 103399. [Google Scholar] [CrossRef]
- Lujambio, A.; Esteller, M. How Epigenetics Can Explain Human Metastasis: A New Role for MicroRNAs. Cell Cycle 2009, 8, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. DNA Methylation and MicroRNA Dysregulation in Cancer. Mol. Oncol. 2012, 6, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Munk, R.; Panda, A.C.; Grammatikakis, I.; Gorospe, M.; Abdelmohsen, K. Senescence-Associated MicroRNAs. Int. Rev. Cell Mol. Biol. 2017, 334, 177–205. [Google Scholar] [PubMed]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Rothbart, S.B.; Strahl, B.D. Interpreting the Language of Histone and DNA Modifications. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2014, 1839, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Nimura, K.; Ura, K.; Kaneda, Y. Histone Methyltransferases: Regulation of Transcription and Contribution to Human Disease. J. Mol. Med. 2010, 88, 1213–1220. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Steger, D.J.; Lefterova, M.I.; Ying, L.; Stonestrom, A.J.; Schupp, M.; Zhuo, D.; Vakoc, A.L.; Kim, J.-E.; Chen, J.; Lazar, M.A.; et al. DOT1L/KMT4 Recruitment and H3K79 Methylation Are Ubiquitously Coupled with Gene Transcription in Mammalian Cells. Mol. Cell. Biol. 2008, 28, 2825–2839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The Interplay of Histone Modifications—Writers That Read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef]
- Kang, W.K.; Florman, J.T.; Araya, A.; Fox, B.W.; Thackeray, A.; Schroeder, F.C.; Walhout, A.J.M.; Alkema, M.J. Vitamin B12 Produced by Gut Bacteria Modulates Cholinergic Signalling. Nat. Cell Biol. 2024, 26, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Kolasinska-Zwierz, P.; Down, T.; Latorre, I.; Liu, T.; Liu, X.S.; Ahringer, J. Differential Chromatin Marking of Introns and Expressed Exons by H3K36me3. Nat. Genet. 2009, 41, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Rapelli, S.; Krepelova, A.; Incarnato, D.; Parlato, C.; Basile, G.; Maldotti, M.; Anselmi, F.; Oliviero, S. Intragenic DNA Methylation Prevents Spurious Transcription Initiation. Nature 2017, 543, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D. DNMT3L Connects Unmethylated Lysine 4 of Histone H3 to de Novo Methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef]
- Henckel, A.; Nakabayashi, K.; Sanz, L.A.; Feil, R.; Hata, K.; Arnaud, P. Histone Methylation Is Mechanistically Linked to DNA Methylation at Imprinting Control Regions in Mammals. Hum. Mol. Genet. 2009, 18, 3375–3383. [Google Scholar] [CrossRef]
- Benevolenskaya, E.V. Histone H3K4 Demethylases Are Essential in Development and Differentiation. Biochem. Cell Biol. 2007, 85, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.M.; Andrews, R.M.; Flicek, P.; Dillon, S.C.; Karaöz, U.; Clelland, G.K.; Wilcox, S.; Beare, D.M.; Fowler, J.C.; Couttet, P. The Landscape of Histone Modifications across 1% of the Human Genome in Five Human Cell Lines. Genome Res. 2007, 17, 691–707. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac Separates Active from Poised Enhancers and Predicts Developmental State. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Pradeepa, M.M.; Grimes, G.R.; Kumar, Y.; Olley, G.; Taylor, G.C.; Schneider, R.; Bickmore, W.A. Histone H3 Globular Domain Acetylation Identifies a New Class of Enhancers. Nat. Genet. 2016, 48, 681–686. [Google Scholar] [CrossRef]
- Fuks, F.; Hurd, P.J.; Deplus, R.; Kouzarides, T. The DNA Methyltransferases Associate with HP1 and the SUV39H1 Histone Methyltransferase. Nucleic Acids Res. 2003, 31, 2305–2312. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.A.; Xu, W.-S. Histone Deacetylase Inhibitors: Potential in Cancer Therapy. J. Cell. Biochem. 2009, 107, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the Protein Deacetylase SIRT6 by Long-Chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Lu, Z.; Xie, Z.; Cheng, Z.; Chen, Y.; Tan, M.; Luo, H.; Zhang, Y.; He, W.; Yang, K. The First Identification of Lysine Malonylation Substrates and Its Regulatory Enzyme. Mol. Cell. Proteom. 2011, 10, M111.012658. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Lee, J.-H.; Lee, H.-Y.; Min, K.-J. Sirtuin Signaling in Cellular Senescence and Aging. BMB Rep. 2019, 52, 24–34. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Paluvai, H.; Di Giorgio, E.; Brancolini, C. The Histone Code of Senescence. Cells 2020, 9, 466. [Google Scholar] [CrossRef]
- Harrington, D.J. Methods for Assessment of Vitamin B12. In Laboratory Assessment of Vitamin Status; Elsevier: Amsterdam, The Netherlands, 2019; pp. 265–299. ISBN 978-0-12-813050-6. [Google Scholar]
- Sobczyńska-Malefora, A.; Delvin, E.; McCaddon, A.; Ahmadi, K.R.; Harrington, D.J. Vitamin B12 Status in Health and Disease: A Critical Review. Diagnosis of Deficiency and Insufficiency—Clinical and Laboratory Pitfalls. Crit. Rev. Clin. Lab. Sci. 2021, 58, 399–429. [Google Scholar] [CrossRef]
- İspir, E.; Serdar, M.A.; Ozgurtas, T.; Gulbahar, O.; Akın, K.O.; Yesildal, F.; Kurt, İ. Comparison of Four Automated Serum Vitamin B12 Assays. Clin. Chem. Lab. Med. 2015, 53, 1205–1213. [Google Scholar] [CrossRef]
- Hannibal, L.; Axhemi, A.; Glushchenko, A.V.; Moreira, E.S.; Brasch, N.E.; Jacobsen, D.W. Accurate Assessment and Identification of Naturally Occurring Cellular Cobalamins. Clin. Chem. Lab. Med. 2008, 46, 1739–1746. [Google Scholar] [CrossRef] [PubMed]
- Green, R.; Allen, L.H.; Bjørke-Monsen, A.-L.; Brito, A.; Guéant, J.-L.; Miller, J.W.; Molloy, A.M.; Nexo, E.; Stabler, S.; Toh, B.-H.; et al. Vitamin B12 Deficiency. Nat. Rev. Dis. Primer 2017, 3, 17040. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Dangat, K.; Kale, A.; Sable, P.; Chavan-Gautam, P.; Joshi, S. Effects of Altered Maternal Folic Acid, Vitamin B12 and Docosahexaenoic Acid on Placental Global DNA Methylation Patterns in Wistar Rats. PLoS ONE 2011, 6, e17706. [Google Scholar] [CrossRef]
- Kumar, K.A.; Lalitha, A.; Reddy, U.; Chandak, G.R.; Sengupta, S.; Raghunath, M. Chronic Maternal Vitamin B12 Restriction Induced Changes in Body Composition & Glucose Metabolism in the Wistar Rat Offspring Are Partly Correctable by Rehabilitation. PLoS ONE 2014, 9, e112991. [Google Scholar]
- Tanwar, V.S.; Ghosh, S.; Sati, S.; Ghose, S.; Kaur, L.; Kumar, K.A.; Shamsudheen, K.V.; Patowary, A.; Singh, M.; Jyothi, V.; et al. Maternal Vitamin B12 Deficiency in Rats Alters DNA Methylation in Metabolically Important Genes in Their Offspring. Mol. Cell. Biochem. 2020, 468, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, K.D.; Allegrucci, C.; Singh, R.; Gardner, D.S.; Sebastian, S.; Bispham, J.; Thurston, A.; Huntley, J.F.; Rees, W.D.; Maloney, C.A.; et al. DNA Methylation, Insulin Resistance, and Blood Pressure in Offspring Determined by Maternal Periconceptional B Vitamin and Methionine Status. Proc. Natl. Acad. Sci. USA 2007, 104, 19351–19356. [Google Scholar] [CrossRef]
- Sharma, G.S.; Kumar, T.; Singh, L.R. N-Homocysteinylation Induces Different Structural and Functional Consequences on Acidic and Basic Proteins. PLoS ONE 2014, 9, e116386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bai, B.; Mei, X.; Wan, C.; Cao, H.; Dan, L.; Wang, S.; Zhang, M.; Wang, Z.; Wu, J.; et al. Elevated H3K79 Homocysteinylation Causes Abnormal Gene Expression during Neural Development and Subsequent Neural Tube Defects. Nat. Commun. 2018, 9, 3436. [Google Scholar] [CrossRef]
- Ghosh, S.; Sinha, J.K.; Khandelwal, N.; Chakravarty, S.; Kumar, A.; Raghunath, M. Increased Stress and Altered Expression of Histone Modifying Enzymes in Brain Are Associated with Aberrant Behaviour in Vitamin B12 Deficient Female Mice. Nutr. Neurosci. 2020, 23, 714–723. [Google Scholar] [CrossRef]
- Suarez-Moreira, E.; Yun, J.; Birch, C.S.; Williams, J.H.H.; McCaddon, A.; Brasch, N.E. Vitamin B12 and Redox Homeostasis: Cob(II)Alamin Reacts with Superoxide at Rates Approaching Superoxide Dismutase (SOD). J. Am. Chem. Soc. 2009, 131, 15078–15079. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Bahnson, E.M.; Wilder, J.; Siletzky, R.; Hagaman, J.; Nickekeit, V.; Hiller, S.; Ayesha, A.; Feng, L.; Levine, J.S.; et al. Oral High Dose Vitamin B12 Decreases Renal Superoxide and Post-Ischemia/Reperfusion Injury in Mice. Redox Biol. 2020, 32, 101504. [Google Scholar] [CrossRef] [PubMed]
- Fowler, B.; Leonard, J.V.; Baumgartner, M.R. Causes of and Diagnostic Approach to Methylmalonic Acidurias. J. Inherit. Metab. Dis. 2008, 31, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Cudré-Cung, H.-P.; Zavadakova, P.; do Vale-Pereira, S.; Remacle, N.; Henry, H.; Ivanisevic, J.; Tavel, D.; Braissant, O.; Ballhausen, D. Ammonium Accumulation Is a Primary Effect of 2-Methylcitrate Exposure in an in Vitro Model for Brain Damage in Methylmalonic Aciduria. Mol. Genet. Metab. 2016, 119, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, L.; Viville, S.; Barton, S.C.; Ishino, F.; Keverne, E.B.; Surani, M.A. Abnormal Maternal Behaviour and Growth Retardation Associated with Loss of the Imprinted Gene Mest. Nat. Genet. 1998, 20, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Sapehia, D.; Mahajan, A.; Singh, P.; Kaur, J. High Dietary Folate and Low Vitamin B12 in Parental Diet Disturbs the Epigenetics of Imprinted Genes MEST and PHLDA2 in Mice Placenta. J. Nutr. Biochem. 2023, 118, 109354. [Google Scholar] [CrossRef] [PubMed]
- De Queiroz, K.B.; Cavalcante-Silva, V.; Lopes, F.L.; Rocha, G.A.; D’Almeida, V.; Coimbra, R.S. Vitamin B12 Is Neuroprotective in Experimental Pneumococcal Meningitis through Modulation of Hippocampal DNA Methylation. J. Neuroinflammation 2020, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Batista, K.S.; Cintra, V.M.; Lucena, P.A.; Manhães-de-Castro, R.; Toscano, A.E.; Costa, L.P.; Queiroz, M.E.; de Andrade, S.M.; Guzman-Quevedo, O.; Aquino, J.d.S. The Role of Vitamin B12 in Viral Infections: A Comprehensive Review of Its Relationship with the Muscle–Gut–Brain Axis and Implications for SARS-CoV-2 Infection. Nutr. Rev. 2022, 80, 561–578. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, F.; Nikbakht, F.; Bahmanpour, S.; Rastegar, K.; Namavar, R. Neuroprotective Effects of NMDA and Group I Metabotropic Glutamate Receptor Antagonists Against Neurodegeneration Induced by Homocysteine in Rat Hippocampus: In Vivo Study. J. Mol. Neurosci. 2013, 50, 551–557. [Google Scholar] [CrossRef]
- Pyrgioti, E.E.; Karakousis, N.D. B12 Levels and Frailty Syndrome. J. Frailty Sarcopenia Falls 2022, 7, 32–37. [Google Scholar] [CrossRef]
- Liu, L.; van Groen, T.; Kadish, I.; Tollefsbol, T.O. DNA Methylation Impacts on Learning and Memory in Aging. Neurobiol. Aging 2009, 30, 549–560. [Google Scholar] [CrossRef]
- Oliveira, A.M.; Hemstedt, T.J.; Bading, H. Rescue of Aging-Associated Decline in Dnmt3a2 Expression Restores Cognitive Abilities. Nat. Neurosci. 2012, 15, 1111–1113. [Google Scholar] [CrossRef]
- Saunderson, E.A.; Spiers, H.; Mifsud, K.R.; Gutierrez-Mecinas, M.; Trollope, A.F.; Shaikh, A.; Mill, J.; Reul, J.M.H.M. Stress-Induced Gene Expression and Behavior Are Controlled by DNA Methylation and Methyl Donor Availability in the Dentate Gyrus. Proc. Natl. Acad. Sci. USA 2016, 113, 4830–4835. [Google Scholar] [CrossRef]
- Loenen, W.A. S-Adenosylmethionine Metabolism and Aging. In Epigenetics of Aging and Longevity; Elsevier: Amsterdam, The Netherlands, 2018; pp. 59–93. [Google Scholar]
- Shen, L.; Ji, H.-F. Associations between Homocysteine, Folic Acid, Vitamin B12 and Alzheimer’s Disease: Insights from Meta-Analyses. J. Alzheimers Dis. 2015, 46, 777–790. [Google Scholar] [CrossRef]
- McCarter, S.J.; Teigen, L.M.; McCarter, A.R.; Benarroch, E.E.; St. Louis, E.K.; Savica, R. Low Vitamin B12 and Parkinson Disease. Mayo Clin. Proc. 2019, 94, 757–762. [Google Scholar] [CrossRef]
- Andrès, E.; Serraj, K.; Zhu, J.; Vermorken, A.J. The Pathophysiology of Elevated Vitamin B12 in Clinical Practice. QJM Int. J. Med. 2013, 106, 505–515. [Google Scholar] [CrossRef]
- Guest, J.; Bilgin, A.; Hokin, B.; Mori, T.A.; Croft, K.D.; Grant, R. Novel Relationships between B12, Folate and Markers of Inflammation, Oxidative Stress and NAD(H) Levels, Systemically and in the CNS of a Healthy Human Cohort. Nutr. Neurosci. 2015, 18, 355–364. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194. [Google Scholar] [CrossRef]
- Tikhonov, S.; Batin, M.; Gladyshev, V.N.; Dmitriev, S.E.; Tyshkovskiy, A. AgeMeta: Quantitative Gene Expression Database of Mammalian Aging. Biochem. Mosc. 2024, 89, 313–321. [Google Scholar] [CrossRef]
- Argan, O.; Ural, D.; Karauzum, K.; Bozyel, S.; Aktaş, M.; Karauzum, I.; Kozdag, G.; Agacdiken Agir, A. Elevated Levels of Vitamin B12 in Chronic Stable Heart Failure: A Marker for Subclinical Liver Damage and Impaired Prognosis. Ther. Clin. Risk Manag. 2018, 14, 1067–1073. [Google Scholar] [CrossRef]
- Mendonça, N.; Jagger, C.; Granic, A.; Martin-Ruiz, C.; Mathers, J.C.; Seal, C.J.; Hill, T.R. Elevated Total Homocysteine in All Participants and Plasma Vitamin B12 Concentrations in Women Are Associated With All-Cause and Cardiovascular Mortality in the Very Old: The Newcastle 85+ Study. J. Gerontol. Ser. A 2018, 73, 1258–1264. [Google Scholar] [CrossRef]
- Sviri, S.; Khalaila, R.; Daher, S.; Bayya, A.; Linton, D.M.; Stav, I.; van Heerden, P.V. Increased Vitamin B12 Levels Are Associated with Mortality in Critically Ill Medical Patients. Clin. Nutr. 2012, 31, 53–59. [Google Scholar] [CrossRef]
- Ogrodnik, M. Cellular Aging beyond Cellular Senescence: Markers of Senescence Prior to Cell Cycle Arrest in Vitro and in Vivo. Aging Cell 2021, 20, e13338. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Wang, Z.; Liu, J.-P. Roles of Telomere Biology in Cell Senescence, Replicative and Chronological Ageing. Cells 2019, 8, 54. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- de Magalhães, J.P. Distinguishing between Driver and Passenger Mechanisms of Aging. Nat. Genet. 2024, 56, 204–211. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; Erusalimsky, J.D.; Campisi, J.; Toussaint, O. Protocols to Detect Senescence-Associated Beta-Galactosidase (SA-Βgal) Activity, a Biomarker of Senescent Cells in Culture and in Vivo. Nat. Protoc. 2009, 4, 1798–1806. [Google Scholar] [CrossRef]
- Lee, B.Y.; Han, J.A.; Im, J.S.; Morrone, A.; Johung, K.; Goodwin, E.C.; Kleijer, W.J.; DiMaio, D.; Hwang, E.S. Senescence-Associated β-Galactosidase Is Lysosomal β-Galactosidase. Aging Cell 2006, 5, 187–195. [Google Scholar] [CrossRef]
- Nobori, T.; Miura, K.; Wu, D.J.; Lois, A.; Takabayashi, K.; Carson, D.A. Deletions of the Cyclin-Dependent Kinase-4 Inhibitor Gene in Multiple Human Cancers. Nature 1994, 368, 753–756. [Google Scholar] [CrossRef]
- Liu, Y.; Sanoff, H.K.; Cho, H.; Burd, C.E.; Torrice, C.; Ibrahim, J.G.; Thomas, N.E.; Sharpless, N.E. Expression of P16 INK4a in Peripheral Blood T-cells Is a Biomarker of Human Aging. Aging Cell 2009, 8, 439–448. [Google Scholar] [CrossRef]
- Sasaki, M.; Kajiya, H.; Ozeki, S.; Okabe, K.; Ikebe, T. Reactive Oxygen Species Promotes Cellular Senescence in Normal Human Epidermal Keratinocytes through Epigenetic Regulation of P16INK4a. Biochem. Biophys. Res. Commun. 2014, 452, 622–628. [Google Scholar] [CrossRef]
- Warfel, N.A.; El-Deiry, W.S. P21WAF1 and Tumourigenesis: 20 Years After. Curr. Opin. Oncol. 2013, 25, 52–58. [Google Scholar] [CrossRef]
- Kamal, M.; Joanisse, S.; Parise, G. Bleomycin-Treated Myoblasts Undergo P21-Associated Cellular Senescence and Have Severely Impaired Differentiation. GeroScience 2023, 46, 1843–1859. [Google Scholar] [CrossRef]
- Stein, G.H.; Drullinger, L.F.; Soulard, A.; Dulić, V. Differential Roles for Cyclin-Dependent Kinase Inhibitors P21 and P16 in the Mechanisms of Senescence and Differentiation in Human Fibroblasts. Mol. Cell. Biol. 1999, 19, 2109–2117. [Google Scholar] [CrossRef]
- Shaw, P.H. The Role of P53 in Cell Cycle Regulation. Pathol.-Res. Pract. 1996, 192, 669–675. [Google Scholar] [CrossRef]
- Sturmlechner, I.; Sine, C.C.; Jeganathan, K.B.; Zhang, C.; Fierro Velasco, R.O.; Baker, D.J.; Li, H.; van Deursen, J.M. Senescent Cells Limit P53 Activity via Multiple Mechanisms to Remain Viable. Nat. Commun. 2022, 13, 3722. [Google Scholar] [CrossRef]
- Beausejour, C.M. Reversal of Human Cellular Senescence: Roles of the P53 and P16 Pathways. EMBO J. 2003, 22, 4212–4222. [Google Scholar] [CrossRef]
- Coppé, J.-P.; Desprez, P.-Y.; Krtolica, A.; Campisi, J. The Senescence-Associated Secretory Phenotype: The Dark Side of Tumor Suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Cuollo, L.; Antonangeli, F.; Santoni, A.; Soriani, A. The Senescence-Associated Secretory Phenotype (SASP) in the Challenging Future of Cancer Therapy and Age-Related Diseases. Biology 2020, 9, 485. [Google Scholar] [CrossRef]
- Polsky, L.R.; Rentscher, K.E.; Carroll, J.E. Stress-Induced Biological Aging: A Review and Guide for Research Priorities. Brain. Behav. Immun. 2022, 104, 97–109. [Google Scholar] [CrossRef]
- Kang, T.-W.; Yevsa, T.; Woller, N.; Hoenicke, L.; Wuestefeld, T.; Dauch, D.; Hohmeyer, A.; Gereke, M.; Rudalska, R.; Potapova, A.; et al. Senescence Surveillance of Pre-Malignant Hepatocytes Limits Liver Cancer Development. Nature 2011, 479, 547–551. [Google Scholar] [CrossRef]
- Acosta, J.C.; O’Loghlen, A.; Banito, A.; Guijarro, M.V.; Augert, A.; Raguz, S.; Fumagalli, M.; Da Costa, M.; Brown, C.; Popov, N.; et al. Chemokine Signaling via the CXCR2 Receptor Reinforces Senescence. Cell 2008, 133, 1006–1018. [Google Scholar] [CrossRef]
- Pribluda, A.; Elyada, E.; Wiener, Z.; Hamza, H.; Goldstein, R.E.; Biton, M.; Burstain, I.; Morgenstern, Y.; Brachya, G.; Billauer, H.; et al. A Senescence-Inflammatory Switch from Cancer-Inhibitory to Cancer-Promoting Mechanism. Cancer Cell 2013, 24, 242–256. [Google Scholar] [CrossRef]
- Eggert, T.; Wolter, K.; Ji, J.; Ma, C.; Yevsa, T.; Klotz, S.; Medina-Echeverz, J.; Longerich, T.; Forgues, M.; Reisinger, F.; et al. Distinct Functions of Senescence-Associated Immune Responses in Liver Tumor Surveillance and Tumor Progression. Cancer Cell 2016, 30, 533–547. [Google Scholar] [CrossRef]
- Canino, C.; Mori, F.; Cambria, A.; Diamantini, A.; Germoni, S.; Alessandrini, G.; Borsellino, G.; Galati, R.; Battistini, L.; Blandino, R.; et al. SASP Mediates Chemoresistance and Tumor-Initiating-Activity of Mesothelioma Cells. Oncogene 2012, 31, 3148–3163. [Google Scholar] [CrossRef]
- Laberge, R.-M.; Awad, P.; Campisi, J.; Desprez, P.-Y. Epithelial-Mesenchymal Transition Induced by Senescent Fibroblasts. Cancer Microenviron. 2012, 5, 39–44. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Däbritz, J.H.M.; Zhao, Z.; Yu, Y.; Dörr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-Associated Reprogramming Promotes Cancer Stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef]
- Nacarelli, T.; Fukumoto, T.; Zundell, J.A.; Fatkhutdinov, N.; Jean, S.; Cadungog, M.G.; Borowsky, M.E.; Zhang, R. NAMPT Inhibition Suppresses Cancer Stem-like Cells Associated with Therapy-Induced Senescence in Ovarian Cancer. Cancer Res. 2020, 80, 890–900. [Google Scholar] [CrossRef]
- Kim, Y.H.; Choi, Y.W.; Lee, J.; Soh, E.Y.; Kim, J.-H.; Park, T.J. Senescent Tumor Cells Lead the Collective Invasion in Thyroid Cancer. Nat. Commun. 2017, 8, 15208. [Google Scholar] [CrossRef]
- Coppé, J.-P.; Kauser, K.; Campisi, J.; Beauséjour, C.M. Secretion of Vascular Endothelial Growth Factor by Primary Human Fibroblasts at Senescence. J. Biol. Chem. 2006, 281, 29568–29574. [Google Scholar] [CrossRef]
- Toste, P.A.; Nguyen, A.H.; Kadera, B.E.; Duong, M.; Wu, N.; Gawlas, I.; Tran, L.M.; Bikhchandani, M.; Li, L.; Patel, S.G.; et al. Chemotherapy-Induced Inflammatory Gene Signature and Protumorigenic Phenotype in Pancreatic CAFs via Stress-Associated MAPK. Mol. Cancer Res. 2016, 14, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Ruhland, M.K.; Loza, A.J.; Capietto, A.-H.; Luo, X.; Knolhoff, B.L.; Flanagan, K.C.; Belt, B.A.; Alspach, E.; Leahy, K.; Luo, J.; et al. Stromal Senescence Establishes an Immunosuppressive Microenvironment That Drives Tumorigenesis. Nat. Commun. 2016, 7, 11762. [Google Scholar] [CrossRef] [PubMed]
- Angelini, P.D.; Fluck, M.F.Z.; Pedersen, K.; Parra-Palau, J.L.; Guiu, M.; Bernadó Morales, C.; Vicario, R.; Luque-García, A.; Navalpotro, N.P.; Giralt, J.; et al. Constitutive HER2 Signaling Promotes Breast Cancer Metastasis through Cellular Senescence. Cancer Res. 2013, 73, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Demaria, M.; O’Leary, M.N.; Chang, J.; Shao, L.; Liu, S.; Alimirah, F.; Koenig, K.; Le, C.; Mitin, N.; Deal, A.M.; et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discov. 2017, 7, 165–176. [Google Scholar] [CrossRef]
- Georgilis, A.; Klotz, S.; Hanley, C.J.; Herranz, N.; Weirich, B.; Morancho, B.; Leote, A.C.; D’Artista, L.; Gallage, S.; Seehawer, M.; et al. PTBP1-Mediated Alternative Splicing Regulates the Inflammatory Secretome and the Pro-Tumorigenic Effects of Senescent Cells. Cancer Cell 2018, 34, 85–102.e9. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.G.; Pant, V.; Li, Q.; Chang, L.L.; Quintás-Cardama, A.; Garza, D.; Tavana, O.; Yang, P.; Manshouri, T.; Li, Y.; et al. P53-Mediated Senescence Impairs the Apoptotic Response to Chemotherapy and Clinical Outcome in Breast Cancer. Cancer Cell 2012, 21, 793–806. [Google Scholar] [CrossRef]
- Sun, Y.; Campisi, J.; Higano, C.; Beer, T.M.; Porter, P.; Coleman, I.; True, L.; Nelson, P.S. Treatment-Induced Damage to the Tumor Microenvironment Promotes Prostate Cancer Therapy Resistance through WNT16B. Nat. Med. 2012, 18, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Takasugi, M.; Yoshida, Y.; Hara, E.; Ohtani, N. The Role of Cellular Senescence and SASP in Tumour Microenvironment. FEBS J. 2023, 290, 1348–1361. [Google Scholar] [CrossRef]
- Carpenter, V.J.; Saleh, T.; Gewirtz, D.A. Senolytics for Cancer Therapy: Is All That Glitters Really Gold? Cancers 2021, 13, 723. [Google Scholar] [CrossRef]
- Khosla, S. Senescent Cells, Senolytics and Tissue Repair: The Devil May Be in the Dosing. Nat. Aging 2023, 3, 139–141. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Lapasset, L.; Milhavet, O.; Prieur, A.; Besnard, E.; Babled, A.; Aït-Hamou, N.; Leschik, J.; Pellestor, F.; Ramirez, J.-M.; De Vos, J.; et al. Rejuvenating Senescent and Centenarian Human Cells by Reprogramming through the Pluripotent State. Genes Dev. 2011, 25, 2248–2253. [Google Scholar] [CrossRef] [PubMed]
- Grigorash, B.B.; van Essen, D.; Liang, G.; Grosse, L.; Emelyanov, A.; Kang, Z.; Korablev, A.; Kanzler, B.; Molina, C.; Lopez, E.; et al. P16High Senescence Restricts Cellular Plasticity during Somatic Cell Reprogramming. Nat. Cell Biol. 2023, 25, 1265–1278. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva, M.; Melendez, E.; Chondronasiou, D.; Pietrocola, F.; Bernad, R.; Caballe, A.; Junza, A.; Capellades, J.; Holguín-Horcajo, A.; Prats, N.; et al. Vitamin B12 Is a Limiting Factor for Induced Cellular Plasticity and Tissue Repair. Nat. Metab. 2023, 5, 1911–1930. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Nuñez, S.; Heard, E.; Narita, M.; Lin, A.W.; Hearn, S.A.; Spector, D.L.; Hannon, G.J.; Lowe, S.W. Rb-Mediated Heterochromatin Formation and Silencing of E2F Target Genes during Cellular Senescence. Cell 2003, 113, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Chandra, T.; Kirschner, K.; Thuret, J.-Y.; Pope, B.D.; Ryba, T.; Newman, S.; Ahmed, K.; Samarajiwa, S.A.; Salama, R.; Carroll, T.; et al. Independence of Repressive Histone Marks and Chromatin Compaction during Senescent Heterochromatic Layer Formation. Mol. Cell 2012, 47, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Chandra, T.; Narita, M. High-Order Chromatin Structure and the Epigenome in SAHFs. Nucleus 2013, 4, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Park, L.K.; Friso, S.; Choi, S.-W. Nutritional Influences on Epigenetics and Age-Related Disease. Proc. Nutr. Soc. 2012, 71, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Sulli, G.; Dobreva, M.; Liontos, M.; Botrugno, O.A.; Gargiulo, G.; dal Zuffo, R.; Matti, V.; d’Ario, G.; Montani, E.; et al. Interplay between Oncogene-Induced DNA Damage Response and Heterochromatin in Senescence and Cancer. Nat. Cell Biol. 2011, 13, 292–302. [Google Scholar] [CrossRef]
- Sidler, C.; Kovalchuk, O.; Kovalchuk, I. Epigenetic Regulation of Cellular Senescence and Aging. Front. Genet. 2017, 8, 138. [Google Scholar] [CrossRef]
- Sarg, B.; Koutzamani, E.; Helliger, W.; Rundquist, I.; Lindner, H.H. Postsynthetic Trimethylation of Histone H4 at Lysine 20 in Mammalian Tissues Is Associated with Aging. J. Biol. Chem. 2002, 277, 39195–39201. [Google Scholar] [CrossRef] [PubMed]
- Ramponi, V.; Richart, L.; Kovatcheva, M.; Stephan-Otto Attolini, C.; Capellades, J.; Lord, A.E.; Yanes, O.; Ficz, G.; Serrano, M. Persister Cancer Cells Are Characterized by H4K20me3 Heterochromatin That Defines a Low Inflammatory Profile. bioRxiv 2024. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Z.; Zhang, L.; Ghosh, S.; Zheng, H.; Zhou, Z. Depleting the Methyltransferase Suv39h1 Improves DNA Repair and Extends Lifespan in a Progeria Mouse Model. Nat. Commun. 2013, 4, 1868. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T. Reversal of the Cellular Phenotype in the Premature Aging Disease Hutchinson-Gilford Progeria Syndrome. Nat. Med. 2005, 11, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant Nuclear Lamin A Leads to Progressive Alterations of Epigenetic Control in Premature Aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef]
- Pusceddu, I.; Herrmann, W.; Kleber, M.E.; Scharnagl, H.; März, W.; Herrmann, M. Telomere Length, Vitamin B12 and Mortality in Persons Undergoing Coronary Angiography: The Ludwigshafen Risk and Cardiovascular Health Study. Aging 2019, 11, 7083–7097. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Liu, D.; Songyang, Z. The Telosome/Shelterin Complex and Its Functions. Genome Biol. 2008, 9, 232. [Google Scholar] [CrossRef]
- Xu, Q.; Parks, C.G.; DeRoo, L.A.; Cawthon, R.M.; Sandler, D.P.; Chen, H. Multivitamin Use and Telomere Length in Women. Am. J. Clin. Nutr. 2009, 89, 1857–1863. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Yang, T.P.; Berry, R.J.; Bailey, L.B. Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate’s Role. Adv. Nutr. 2012, 3, 21–38. [Google Scholar] [CrossRef]
- Rzepka, Z.; Rok, J.; Kowalska, J.; Banach, K.; Wrześniok, D. Cobalamin Deficiency May Induce Astrosenescence—An In Vitro Study. Cells 2022, 11, 3408. [Google Scholar] [CrossRef]
- Kwan, D.N.; Rocha, J.T.Q.; Niero-Melo, L.; Custódio, M.A.; Oliveira, C.C. P53 and P21 Expression in Bone Marrow Clots of Megaloblastic Anemia Patients. Int. J. Clin. Exp. Pathol. 2020, 13, 1829–1833. [Google Scholar] [PubMed]
- Bhanothu, V.; Venkatesan, V.; Kondapi, A.K.; Ajumeera, R. Restrictions and Supplementations Effects of Vitamins B6, B9 and B12 on Growth, Vasculogenesis and Senescence of BG01V Human Embryonic Stem Cell Derived Embryoid Bodies. Nutr. Clin. Métab. 2021, 35, 297–316. [Google Scholar] [CrossRef]
- Praveen, G.; Sivaprasad, M.; Reddy, G.B. Chapter Eleven—Telomere Length and Vitamin B12. In Vitamins and Hormones; Litwack, G., Ed.; Vitamin B12; Academic Press: Cambridge, MA, USA, 2022; Volume 119, pp. 299–324. [Google Scholar]
- Zhang, R.; Bons, J.; Scheidemantle, G.; Liu, X.; Bielska, O.; Carrico, C.; Rose, J.; Heckenbach, I.; Scheibye-Knudsen, M.; Schilling, B.; et al. Histone Malonylation Is Regulated by SIRT5 and KAT2A. iScience 2023, 26, 106193. [Google Scholar] [CrossRef] [PubMed]
- Ghemrawi, R.; Pooya, S.; Lorentz, S.; Gauchotte, G.; Arnold, C.; Gueant, J.-L.; Battaglia-Hsu, S.-F. Decreased Vitamin B12 Availability Induces ER Stress through Impaired SIRT1-Deacetylation of HSF1. Cell Death Dis. 2013, 4, e553. [Google Scholar] [CrossRef] [PubMed]
- Cuyàs, E.; Verdura, S.; Llorach-Parés, L.; Fernández-Arroyo, S.; Joven, J.; Martin-Castillo, B.; Bosch-Barrera, J.; Brunet, J.; Nonell-Canals, A.; Sanchez-Martinez, M.; et al. Metformin Is a Direct SIRT1-Activating Compound: Computational Modeling and Experimental Validation. Front. Endocrinol. 2018, 9, 657. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-J.; Defossez, P.-A.; Guarente, L. Requirement of NAD and SIR2 for Life-Span Extension by Calorie Restriction in Saccharomyces cerevisiae. Science 2000, 289, 2126–2128. [Google Scholar] [CrossRef]
- Gomes, A.P.; Ilter, D.; Low, V.; Endress, J.E.; Fernández-García, J.; Rosenzweig, A.; Schild, T.; Broekaert, D.; Ahmed, A.; Planque, M.; et al. Age-Induced Accumulation of Methylmalonic Acid Promotes Tumour Progression. Nature 2020, 585, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Fu, Q.; Sun, Y.; Li, Q. Epigenetic Clock: A Promising Biomarker and Practical Tool in Aging. Ageing Res. Rev. 2022, 81, 101743. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Hayano, M.; Griffin, P.T.; Amorim, J.A.; Bonkowski, M.S.; Apostolides, J.K.; Salfati, E.L.; Blanchette, M.; Munding, E.M.; Bhakta, M. Loss of Epigenetic Information as a Cause of Mammalian Aging. Cell 2023, 186, 305–326. [Google Scholar] [CrossRef]
- Sen, P.; Donahue, G.; Li, C.; Egervari, G.; Yang, N.; Lan, Y.; Robertson, N.; Shah, P.P.; Kerkhoven, E.; Schultz, D.C. Spurious Intragenic Transcription Is a Feature of Mammalian Cellular Senescence and Tissue Aging. Nat. Aging 2023, 3, 402–417. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Nemirovsky, L.E.; Klimenko, V.V.; Vasiliev, V.K.; Belozersky, A.N. The 5-Methylcytosine in DNA of Rats. Gerontology 1973, 19, 138–152. [Google Scholar] [CrossRef]
- Wilson, V.L.; Smith, R.A.; Ma, S.; Cutler, R.G. Genomic 5-Methyldeoxycytidine Decreases with Age. J. Biol. Chem. 1987, 262, 9948–9951. [Google Scholar] [CrossRef]
- Drinkwater, R.D.; Blake, T.J.; Morley, A.A.; Turner, D.R. Human Lymphocytes Aged in Vivo Have Reduced Levels of Methylation in Transcriptionally Active and Inactive DNA. Mutat. Res. 1989, 219, 29–37. [Google Scholar] [CrossRef]
- Fernàndez-Roig, S.; Lai, S.-C.; Murphy, M.M.; Fernandez-Ballart, J.; Quadros, E.V. Vitamin B12 Deficiency in the Brain Leads to DNA Hypomethylation in the TCblR/CD320 Knockout Mouse. Nutr. Metab. 2012, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Amenyah, S.D.; Hughes, C.F.; Ward, M.; Rosborough, S.; Deane, J.; Thursby, S.-J.; Walsh, C.P.; Kok, D.E.; Strain, J.J.; McNulty, H.; et al. Influence of Nutrients Involved in One-Carbon Metabolism on DNA Methylation in Adults—A Systematic Review and Meta-Analysis. Nutr. Rev. 2020, 78, 647–666. [Google Scholar] [CrossRef]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.H.; Erbel, R.; Mühleisen, T.W.; et al. Aging of Blood Can Be Tracked by DNA Methylation Changes at Just Three CpG Sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Hübner, U.; Bodis, M.; Graeber, S.; Geisel, J. Effect of Adding B-Vitamins to Vitamin D and Calcium Supplementation on CpG Methylation of Epigenetic Aging Markers. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 411–417. [Google Scholar] [CrossRef]
- Sae-Lee, C.; Corsi, S.; Barrow, T.M.; Kuhnle, G.G.C.; Bollati, V.; Mathers, J.C.; Byun, H. Dietary Intervention Modifies DNA Methylation Age Assessed by the Epigenetic Clock. Mol. Nutr. Food Res. 2018, 62, 1800092. [Google Scholar] [CrossRef]
- Lee, Y.-T.; Savini, M.; Chen, T.; Yang, J.; Zhao, Q.; Ding, L.; Gao, S.M.; Senturk, M.; Sowa, J.N.; Wang, J.D.; et al. Mitochondrial GTP Metabolism Controls Reproductive Aging in C. Elegans. Dev. Cell 2023, 58, 2718–2731.e7. [Google Scholar] [CrossRef] [PubMed]
- Monasso, G.S.; Küpers, L.K.; Jaddoe, V.W.V.; Heil, S.G.; Felix, J.F. Associations of Circulating Folate, Vitamin B12 and Homocysteine Concentrations in Early Pregnancy and Cord Blood with Epigenetic Gestational Age: The Generation R Study. Clin. Epigenet. 2021, 13, 95. [Google Scholar] [CrossRef]
- Hirata, T.; Arai, Y.; Yuasa, S.; Abe, Y.; Takayama, M.; Sasaki, T.; Kunitomi, A.; Inagaki, H.; Endo, M.; Morinaga, J.; et al. Associations of Cardiovascular Biomarkers and Plasma Albumin with Exceptional Survival to the Highest Ages. Nat. Commun. 2020, 11, 3820. [Google Scholar] [CrossRef]
- Bogdanova, D.A.; Kolosova, E.D.; Pukhalskaia, T.V.; Levchuk, K.A.; Demidov, O.N.; Belotserkovskaya, E.V. The Differential Effect of Senolytics on SASP Cytokine Secretion and Regulation of EMT by CAFs. Int. J. Mol. Sci. 2024, 25, 4031. [Google Scholar] [CrossRef]
- Ortiz-Montero, P.; Londoño-Vallejo, A.; Vernot, J.-P. Senescence-Associated IL-6 and IL-8 Cytokines Induce a Self- and Cross-Reinforced Senescence/Inflammatory Milieu Strengthening Tumorigenic Capabilities in the MCF-7 Breast Cancer Cell Line. Cell Commun. Signal. 2017, 15, 17. [Google Scholar] [CrossRef]
- Christen, W.G.; Cook, N.R.; Van Denburgh, M.; Zaharris, E.; Albert, C.M.; Manson, J.E. Effect of Combined Treatment With Folic Acid, Vitamin B 6, and Vitamin B12 on Plasma Biomarkers of Inflammation and Endothelial Dysfunction in Women. J. Am. Heart Assoc. 2018, 7, e008517. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, C. The Return of the Scarlet Pimpernel: Cobalamin in Inflammation II—Cobalamins Can Both Selectively Promote All Three Nitric Oxide Synthases (NOS), Particularly INOS and ENOS, and, as Needed, Selectively Inhibit INOS and NNOS. J. Nutr. Environ. Med. 2007, 16, 181–211. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Wang, M.-Y.; Lin, M.-C.; Lin, P.-T. Associations between Vitamin B-12 Status and Oxidative Stress and Inflammation in Diabetic Vegetarians and Omnivores. Nutrients 2016, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Gubernatorova, E.O.; Polinova, A.I.; Petropavlovskiy, M.M.; Namakanova, O.A.; Medvedovskaya, A.D.; Zvartsev, R.V.; Telegin, G.B.; Drutskaya, M.S.; Nedospasov, S.A. Dual Role of TNF and LTα in Carcinogenesis as Implicated by Studies in Mice. Cancers 2021, 13, 1775. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, S.P.; Quadros, E.V.; Regec, A. Transcobalamin II. Chem. Biochem. Vitam. B 1999, 12, 441–473. [Google Scholar]
- Scalabrino, G.; Carpo, M.; Bamonti, F.; Pizzinelli, S.; D’Avino, C.; Bresolin, N.; Meucci, G.; Martinelli, V.; Comi, G.C.; Peracchi, M. High Tumor Necrosis Factor-α in Levels in Cerebrospinal Fluid of Cobalamin-deficient Patients. Ann. Neurol. 2004, 56, 886–890. [Google Scholar] [CrossRef] [PubMed]
- Kalra, S.; Ahuja, R.; Mutti, E.; Veber, D.; Seetharam, S.; Scalabrino, G.; Seetharam, B. Cobalamin-Mediated Regulation of Transcobalamin Receptor Levels in Rat Organs. Arch. Biochem. Biophys. 2007, 463, 128–132. [Google Scholar] [CrossRef]
- Veber, D.; Mutti, E.; Tacchini, L.; Gammella, E.; Tredici, G.; Scalabrino, G. Indirect Down-regulation of Nuclear NF-κB Levels by Cobalamin in the Spinal Cord and Liver of the Rat. J. Neurosci. Res. 2008, 86, 1380–1387. [Google Scholar] [CrossRef]
- Homann, L.; Rentschler, M.; Brenner, E.; Böhm, K.; Röcken, M.; Wieder, T. IFN-γ and TNF Induce Senescence and a Distinct Senescence-Associated Secretory Phenotype in Melanoma. Cells 2022, 11, 1514. [Google Scholar] [CrossRef] [PubMed]
- Kandhaya-Pillai, R.; Yang, X.; Tchkonia, T.; Martin, G.M.; Kirkland, J.L.; Oshima, J. TNF -α/ IFN -γ Synergy Amplifies Senescence-associated Inflammation and SARS-CoV -2 Receptor Expression via Hyper-activated JAK / STAT1. Aging Cell 2022, 21, e13646. [Google Scholar] [CrossRef] [PubMed]
- Michelucci, A.; Cordes, T.; Ghelfi, J.; Pailot, A.; Reiling, N.; Goldmann, O.; Binz, T.; Wegner, A.; Tallam, A.; Rausell, A.; et al. Immune-Responsive Gene 1 Protein Links Metabolism to Immunity by Catalyzing Itaconic Acid Production. Proc. Natl. Acad. Sci. USA 2013, 110, 7820–7825. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Campanello, G.C.; Flicker, D.; Grabarek, Z.; Hu, J.; Luo, C.; Banerjee, R.; Mootha, V.K. The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B12. Cell 2017, 171, 771–782.e11. [Google Scholar] [CrossRef] [PubMed]
- Ruetz, M.; Campanello, G.C.; Purchal, M.; Shen, H.; McDevitt, L.; Gouda, H.; Wakabayashi, S.; Zhu, J.; Rubin, E.J.; Warncke, K.; et al. Itaconyl-CoA Forms a Stable Biradical in Methylmalonyl-CoA Mutase and Derails Its Activity and Repair. Science 2019, 366, 589–593. [Google Scholar] [CrossRef]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging: An Evolutionary Perspective on Immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, S.; Zhao, L.; Cheng, P.; Liu, J.; Guo, F.; Xiao, J.; Zhu, W.; Chen, A. Aging Relevant Metabolite Itaconate Inhibits Inflammatory Bone Loss. Front. Endocrinol. 2022, 13, 885879. [Google Scholar] [CrossRef] [PubMed]
- Martucci, M.; Conte, M.; Bucci, L.; Giampieri, E.; Fabbri, C.; Palmas, M.; Izzi, M.; Salvioli, S.; Zambrini, A.; Orsi, C.; et al. Twelve-Week Daily Consumption of Ad Hoc Fortified Milk with ω-3, D, and Group B Vitamins Has a Positive Impact on Inflammaging Parameters: A Randomized Cross-Over Trial. Nutrients 2020, 12, 3580. [Google Scholar] [CrossRef]
- Ghosh, T.S. Toward an Improved Definition of a Healthy Microbiome for Healthy Aging. Nat. Aging 2022, 2, 1054–1069. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut Microbiota Composition Correlates with Diet and Health in the Elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, H.; Lu, W.; Wu, T.; Yuan, W.; Zhu, J.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. Human Gut Microbiome Aging Clocks Based on Taxonomic and Functional Signatures through Multi-View Learning. Gut Microbes 2022, 14, 2025016. [Google Scholar] [CrossRef]
- Berlin, H.; Berlin, R.; Brante, G. Oral treatment of pernicious anemia with high doses of vitamin b12 without intrinsic factor. Acta Med. Scand. 1968, 184, 247–258. [Google Scholar] [CrossRef]
- Kuzminski, A.M.; Del Giacco, E.J.; Allen, R.H.; Stabler, S.P.; Lindenbaum, J. Effective Treatment of Cobalamin Deficiency With Oral Cobalamin. Blood 1998, 92, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Seetharam, B.; Alpers, D.H. Absorption and Transport of Cobalamin (Vitamin B12). Annu. Rev. Nutr. 1982, 2, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.H.; Stabler, S.P. Identification and Quantitation of Cobalamin and Cobalamin Analogues in Human Feces. Am. J. Clin. Nutr. 2008, 87, 1324–1335. [Google Scholar] [CrossRef]
- Bárcena, C.; Valdés-Mas, R.; Mayoral, P.; Garabaya, C.; Durand, S.; Rodríguez, F.; Fernández-García, M.T.; Salazar, N.; Nogacka, A.M.; Garatachea, N.; et al. Healthspan and Lifespan Extension by Fecal Microbiota Transplantation into Progeroid Mice. Nat. Med. 2019, 25, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.; Romano, S.; Ansorge, R.; Aboelnour, A.; Le Gall, G.; Savva, G.M.; Pontifex, M.G.; Telatin, A.; Baker, D.; Jones, E.; et al. Fecal Microbiota Transfer between Young and Aged Mice Reverses Hallmarks of the Aging Gut, Eye, and Brain. Microbiome 2022, 10, 68. [Google Scholar] [CrossRef]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef]
- Sato, Y.; Atarashi, K.; Plichta, D.R.; Arai, Y.; Sasajima, S.; Kearney, S.M.; Suda, W.; Takeshita, K.; Sasaki, T.; Okamoto, S.; et al. Novel Bile Acid Biosynthetic Pathways Are Enriched in the Microbiome of Centenarians. Nature 2021, 599, 458–464. [Google Scholar] [CrossRef]
- Kawamoto, S.; Uemura, K.; Hori, N.; Takayasu, L.; Konishi, Y.; Katoh, K.; Matsumoto, T.; Suzuki, M.; Sakai, Y.; Matsudaira, T.; et al. Bacterial Induction of B Cell Senescence Promotes Age-Related Changes in the Gut Microbiota. Nat. Cell Biol. 2023, 25, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.S.; Gubbi, S.; Barzilai, N. Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab. 2020, 32, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Cuyàs, E.; Fernández-Arroyo, S.; Verdura, S.; García, R.Á.-F.; Stursa, J.; Werner, L.; Blanco-González, E.; Montes-Bayón, M.; Joven, J.; Viollet, B.; et al. Metformin Regulates Global DNA Methylation via Mitochondrial One-Carbon Metabolism. Oncogene 2018, 37, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Aroda, V.R.; Edelstein, S.L.; Goldberg, R.B.; Knowler, W.C.; Marcovina, S.M.; Orchard, T.J.; Bray, G.A.; Schade, D.S.; Temprosa, M.G.; White, N.H.; et al. Long-Term Metformin Use and Vitamin B12 Deficiency in the Diabetes Prevention Program Outcomes Study. J. Clin. Endocrinol. Metab. 2016, 101, 1754–1761. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Verheijen, B.M.; Navapanich, Z.; McGann, E.G.; Shemtov, S.; Lai, G.-J.; Arora, P.; Towheed, A.; Haroon, S.; Holczbauer, A.; et al. Evolutionary Conservation of the Fidelity of Transcription. Nat. Commun. 2023, 14, 1547. [Google Scholar] [CrossRef] [PubMed]
- Debès, C.; Papadakis, A.; Grönke, S.; Karalay, Ö.; Tain, L.S.; Mizi, A.; Nakamura, S.; Hahn, O.; Weigelt, C.; Josipovic, N.; et al. Ageing-Associated Changes in Transcriptional Elongation Influence Longevity. Nature 2023, 616, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Mohsin, M.; Iqrar, S.; Manzoor, O.; Siddiqi, T.O.; Ahmad, A. Live Cell Imaging of Vitamin B12 Dynamics by Genetically Encoded Fluorescent Nanosensor. Sens. Actuators B Chem. 2018, 257, 866–874. [Google Scholar] [CrossRef]
- Soleja, N.; Agrawal, N.; Nazir, R.; Ahmad, M.; Mohsin, M. Enhanced Sensitivity and Detection Range of FRET-Based Vitamin B12 Nanosensor. 3 Biotech 2020, 10, 87. [Google Scholar] [CrossRef]



| Modification | Role | Reference |
|---|---|---|
| H3K9me2 H3K27me2 H3K9me3 H3K27me3 H3K79me3 H4K20me3 H2A/H4R3me2 | Repression of gene expression | [83,84,90] |
| H3K4me1 H3K4me3 H2BK5me1 H3K9me1 H3K27me1 H4K20me1 H3K79me1 H3K79me2 H3K9ac H3K14ac H3K27ac H3K63ac H3K122ac | Increase in gene expression | [83,84,91,92,93,94] |
| H3K36me3 | Prevention of transcription initiation outside promoters | [88] |
| H3K4 methylation | Prevention of DNA methylation | [89] |
| Analyte | Reference Interval | B12 Deficiency | Advantages and Disadvantages |
|---|---|---|---|
| Total B12, pM | 200–600 | <148 | Requires digestion of proteins |
| Holotranscobalamin, pM | 40–100 | <35 | Highly automated |
| Homocysteine, μM | 8–15 | >15 | Several confounders: B9 and B6 levels, thyroid function, glomerular filtration rate, sex, and age |
| 2-methylmalonic acid, nM | 0.04–0.37 | >0.37 | Results may be unreliable in cases of kidney deficiency or infection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simonenko, S.Y.; Bogdanova, D.A.; Kuldyushev, N.A. Emerging Roles of Vitamin B12 in Aging and Inflammation. Int. J. Mol. Sci. 2024, 25, 5044. https://doi.org/10.3390/ijms25095044
Simonenko SY, Bogdanova DA, Kuldyushev NA. Emerging Roles of Vitamin B12 in Aging and Inflammation. International Journal of Molecular Sciences. 2024; 25(9):5044. https://doi.org/10.3390/ijms25095044
Chicago/Turabian StyleSimonenko, Sergey Yu., Daria A. Bogdanova, and Nikita A. Kuldyushev. 2024. "Emerging Roles of Vitamin B12 in Aging and Inflammation" International Journal of Molecular Sciences 25, no. 9: 5044. https://doi.org/10.3390/ijms25095044





