The Transcription Factor MiMYB8 Suppresses Peel Coloration in Postharvest ‘Guifei’ Mango in Response to High Concentration of Exogenous Ethylene by Negatively Modulating MiPAL1
Abstract
:1. Introduction
2. Results
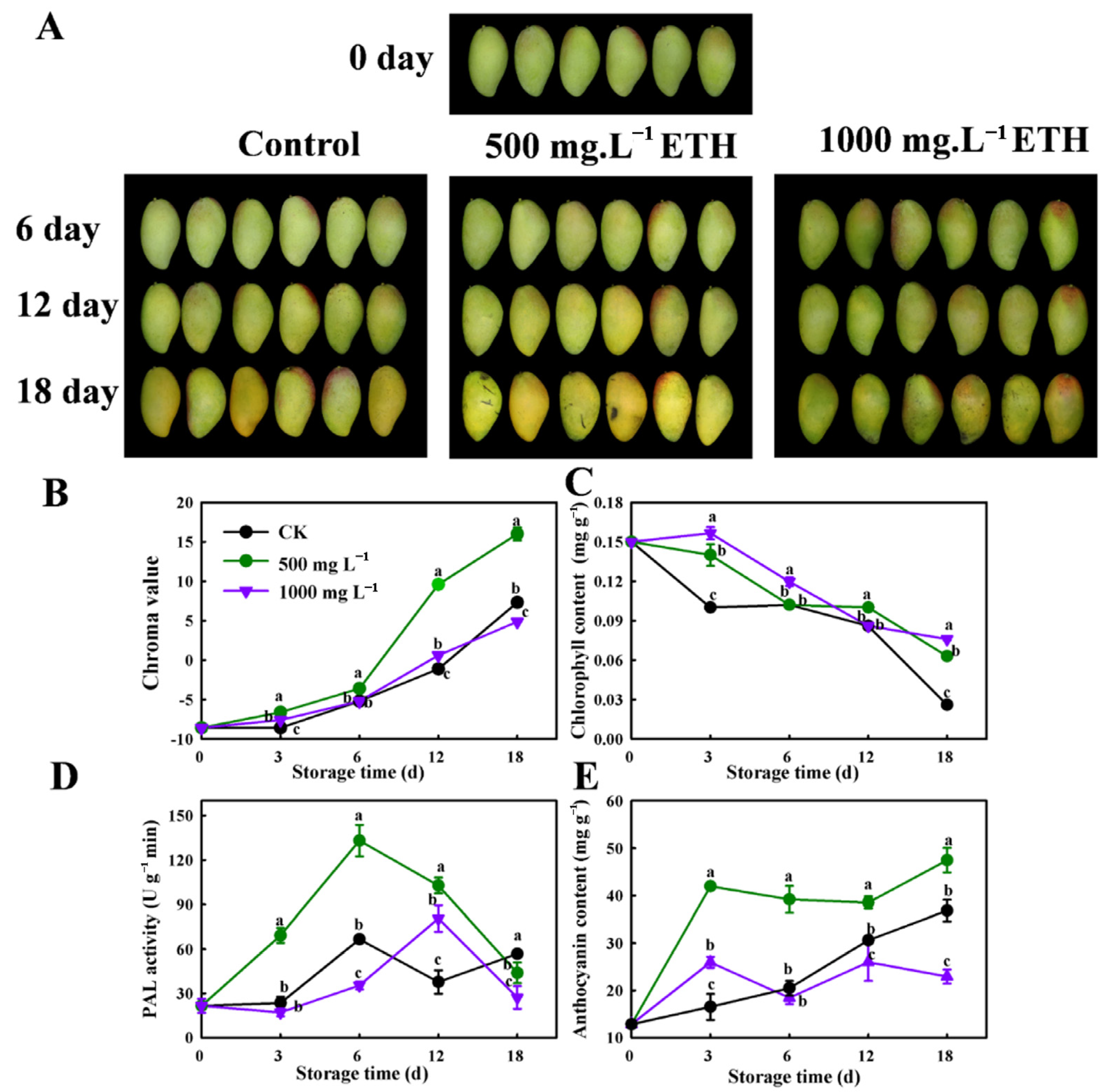
2.1. The Effect of Exogenous Ethylene (ETH) on Peel Coloration in Mango Fruit
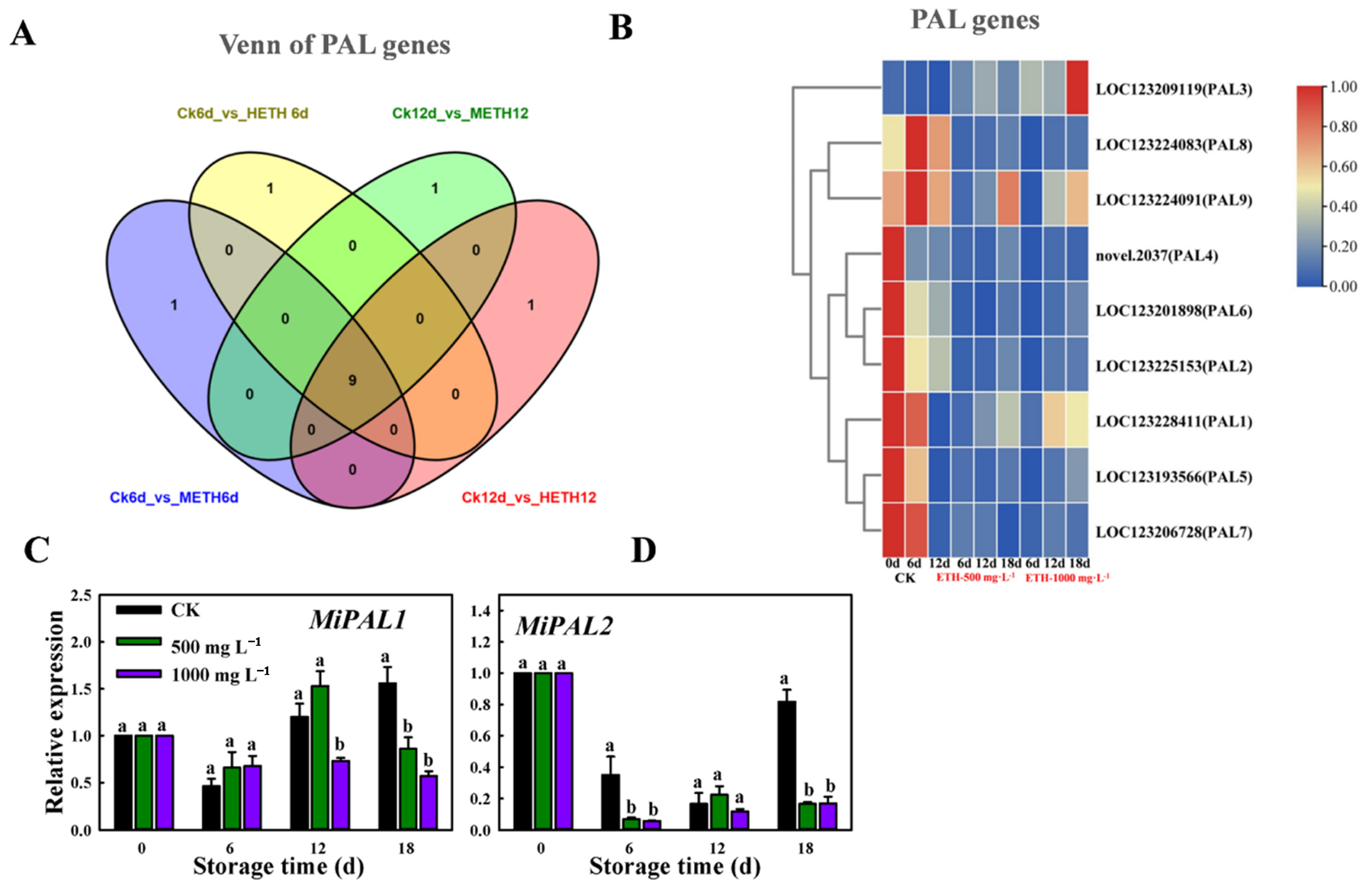
2.2. Transcriptomic Expression Analysis of MiPAL in Mango Fruit
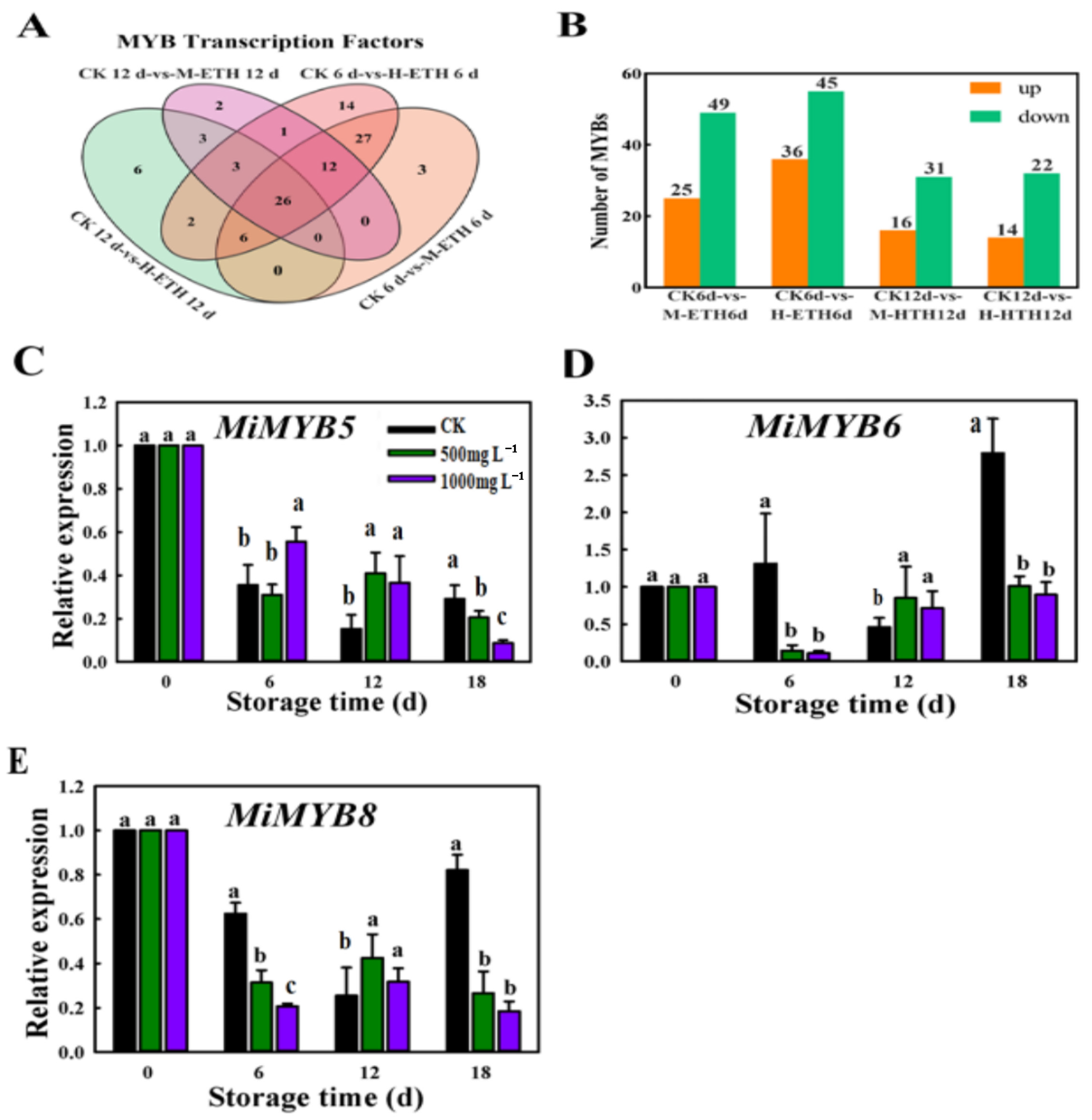
2.3. Transcriptomic Expression Analysis of MiMYB in Mango Fruit
2.4. Phylogenetic Analysis and Subcellular Localization of MiMYB8 Gene
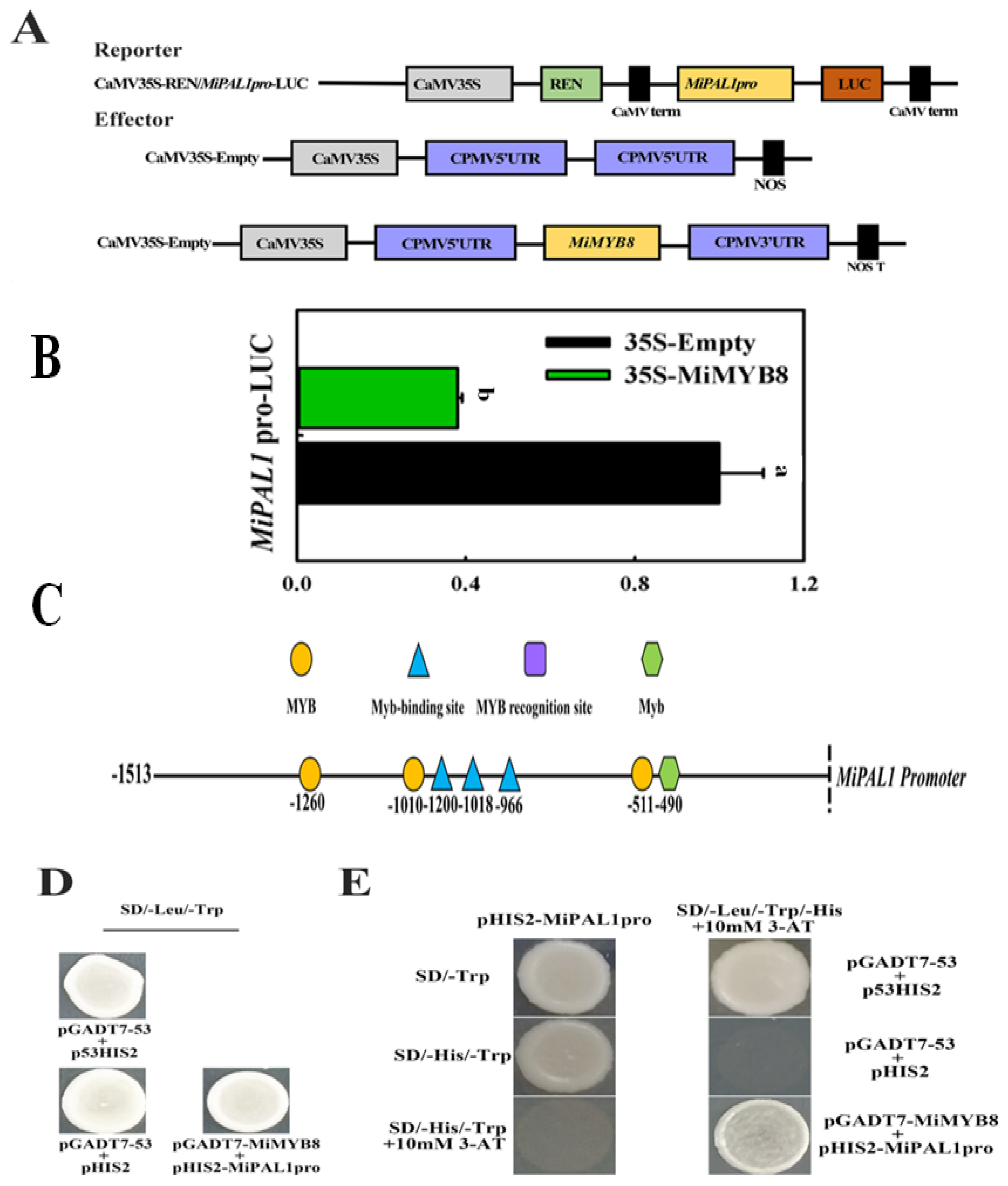
2.5. Interaction of MiMYB8 and MiPAL1 Genes
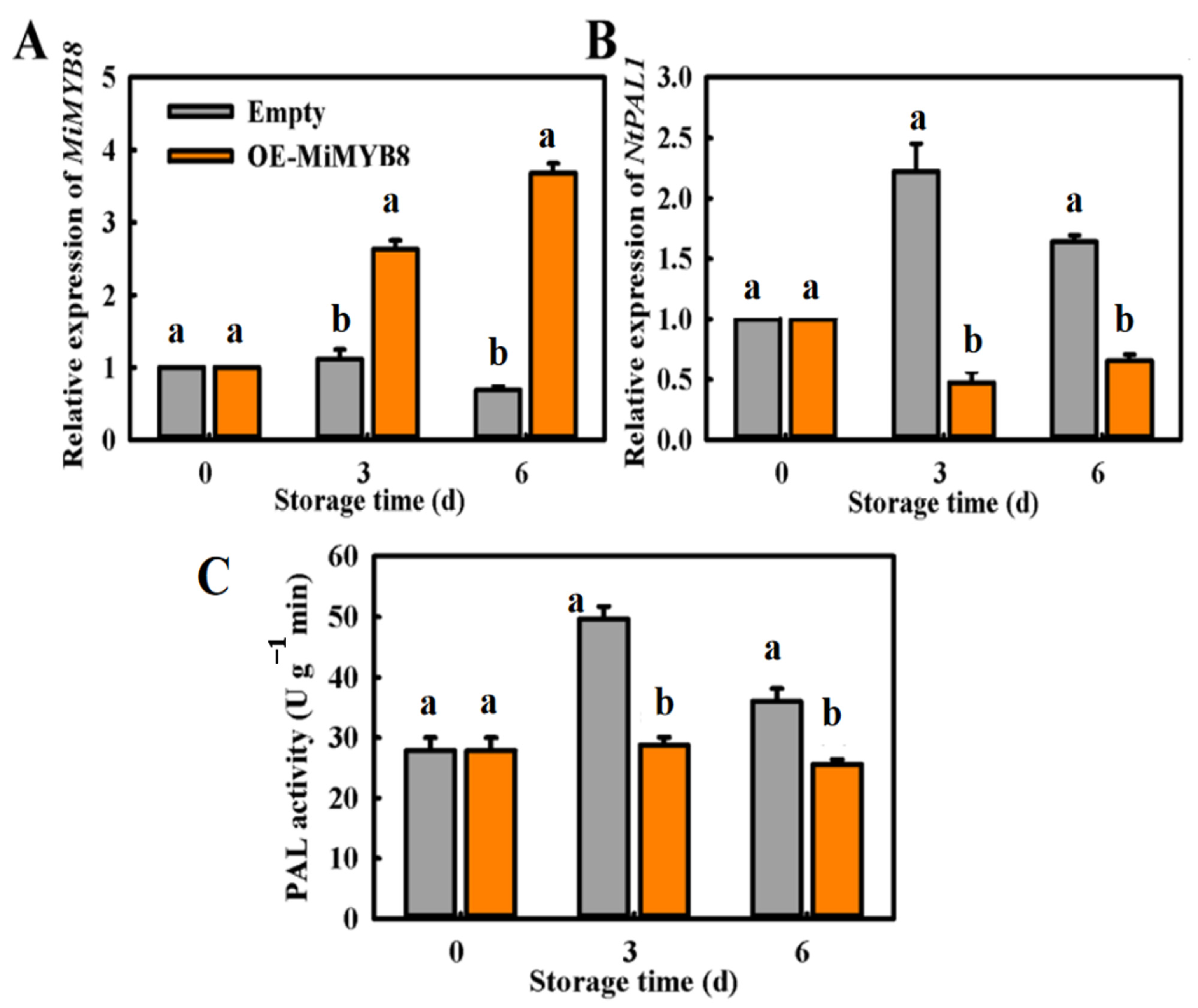
2.6. Transient Overexpression (OE) of MiMYB8 in Tobacco Leaf
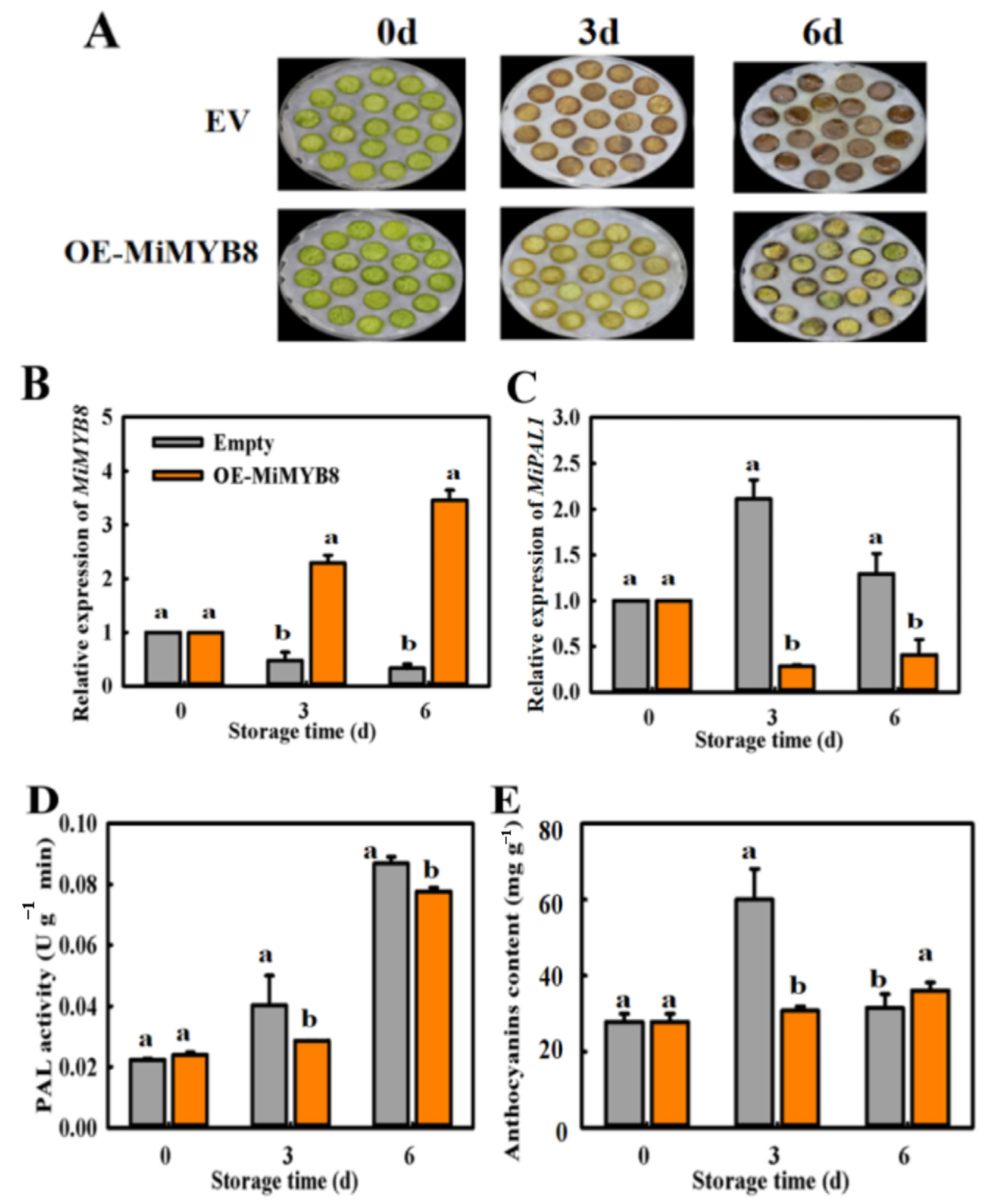
2.7. Transient Overexpression (OE) of MiMYB8 in Mango Fruit
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. Measurement of Peel Color Characteristics of Mango Fruit
4.3. Determination of PAL Enzyme Activity
4.4. Analysis of Differentially Expressed Genes (DEGs)
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Gene Cloning and Promoter Analysis
4.7. Subcellular Localization Analysis
4.8. MiPAL1 Promoter Amplification and Yeast One-Hybrid (Y1H) Assay
4.9. Dual-Luciferase Reporter (DLR) Assays
4.10. Transient Overexpression (OE) of MiMYB8 in Tobacco Leaf
4.11. Transient Overexpression (OE) of MiMYB8 Gene in Mango Fruit
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Y.L.; Fan, L.Q.; He, J.X.; Wang, Z.K.; Yin, Y.P.; Cheng, Y.L.; Li, Z.G. Anthocyanins contribute to fruit defense against postharvest green mold. Postharvest Biol. Technol. 2021, 181, 111661. [Google Scholar] [CrossRef]
- Yan, Y.F.; Pico, J.; Gerbrandt, E.M.; Dossett, M.; Castellarin, S.D. Comprehensive anthocyanin and flavonol profiling and frui surface color of 20 blueberry genotypes during postharvest storage. Postharvest Biol. Technol. 2023, 199, 112274. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, Y.; Zhang, G.L.; Wang, Y.X.; Huang, J.Y.; Feng, Y.W.; Li, Y.; Jiang, J.; Zhang, Y.J. Overexpression of a BR inactivating enzyme gene GhPAG1 impacts eggplant fruit development and anthocyanin accumulation mainly by altering hormone homeostasis. Plant Sci. 2024, 341, 112014. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.L.; Wang, W.J.; Li, T.; Chen, O.; Yao, S.X.; Deng, L.L.; Zeng, K.F. Genome-wide identification of the CsPAL gene family and functional analysis for strengthening green mold resistance in citrus fruit. Postharvest Biol. Technol. 2023, 196, 112178. [Google Scholar] [CrossRef]
- Mingkun Chen, M.K.; Cao, X.Y.; Huang, Y.X.; Zou, W.T.; Liang, X.L.; Yang, Y.; Wang, Y.; Wei, J.; Li, H.H. MpbZIP9 transcription factor regulates biosynthesis of anthocyanin in Malus ‘Pinkspire’ fruit in response to ABA. Plant Sci. 2024, 342, 112038. [Google Scholar] [CrossRef]
- Li, Y.; Xing, M.H.; Yang, Q.; Wang, Y.; Jiang, J.; Zhao, Y.K.; Zhao, X.M.; Shen, A.M.; Feng, Y.W.; Zhao, X.J.; et al. SmCIP7, a COP1 interactive protein, positively regulates anthocyanin accumulation and fruit size in eggplant. Int. J. Biol. Macromol. 2023, 234, 123729. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Ji, N.N.; Zuo, X.X.; Hou, Y.Y.; Zhang, J.L.; Zou, Y.Y.; Jin, P.; Zheng, Y.H. PpMYB308 is involved in Pichia guilliermondii-induced disease resistance against Rhizopus rot by activating the phenylpropanoid pathway in peach fruit. Postharvest Biol. Technol. 2023, 195, 112115. [Google Scholar] [CrossRef]
- Jiang, S.H.; Sun, Q.G.; Zhang, T.L.; Liu, W.J.; Wang, N.; Chen, X.S. MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4-like in apple fruit. J. Plant Physiol. 2021, 257, 153353. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.Y.; Dong, Q.L.; Wu, J.H.; Li, H.; Luan, H.A.; Jia, P.; Zhang, X.M.; Guo, S.P.; Yang, M.S.; Qi, G.H. Genome-wide analysis of the R2R3-MYB transcription factor gene family expressed in Juglans regia under abiotic and biotic stresses. Ind. Crops Prod. 2023, 198, 116709. [Google Scholar] [CrossRef]
- Feng, C.; Zhang, X.; Wang, Y.T.; Zhou, X.; Wang, Y.Y.; Sun, Y.T.; Xiao, Y.Q.; Du, B.Y.; Li, T.H. Effects of water-saving ridging and film-covering cultivation methods on fruit anthocyanin biosynthesis in apple fruit. Sci. Hortic. 2023, 321, 112316. [Google Scholar] [CrossRef]
- Wang, Y.G.; Zhou, L.J.; Wang, Y.X.; Geng, Z.Q.; Ding, B.Q.; Jiang, J.F.; Chen, S.M.; Chen, F.D. An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum. Sci. Hortic. 2022, 293, 110674. [Google Scholar] [CrossRef]
- Duan, S.C.; Eom, S.H. Regulation of anthocyanin and lignin contents in postharvest ‘Fuji’ apple irradiated with UV-B. Sci. Hortic. 2023, 322, 112428. [Google Scholar] [CrossRef]
- Zhang, B.B.; Wang, N.; Chen, X.X.; Wang, X.J.; Zhang, Y.Y.; Guo, S.L.; Yu, M.L.; Ma, R.J. Effect of fruit debagging before harvest on the accumulation of anthocyanins in peach (Prunus persica L.) fruit based on transcriptome analysis. Sci. Hortic. 2023, 308, 111582. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- Figueroa, N.E.; Gatica-Melendez, C.; Figueroa, C.R. Ethylene application at the immature stage of Fragaria chiloensis fruit represses the anthocyanin biosynthesis with a concomitant accumulation of lignin. Food Chem. 2021, 358, 129913. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, S.Z.; Ma, H.P.; Duan, X.J.; Gao, S.X.; Zhou, X.J.; Cheng, Y.W. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef]
- Li, Y.F.; Zhang, J.L.; Ji, N.N.; Zuo, X.X.; Ru, X.Y.; Jin, P.; Wang, K.T.; Zheng, Y.H. PpMYB44 positively affects salicylic acid biosynthesis in Pichia guilliermondii-induced peach fruit resistance against Rhizopus stolonifer. Postharvest Biol. Technol. 2023, 202, 112395. [Google Scholar] [CrossRef]
- Xie, S.; Lei, Y.; Chen, H.; Li, J.; Chen, H.; Zhang, Z. R2R3-MYB transcription factors regulate anthocyanin biosynthesis in grapevine vegetative tissues. Front. Plant Sci. 2020, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Díaz, J.R.; Pérez-Díaz, J.; Madrid-Espinoza, J.; González-Villanueva, E.; Moreno, Y.; Ruiz-Lara, S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016, 90, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, E.; Matus, J.T.; Finezzo, L.; Zenoni, S.; Loyola, R.; Guzzo, F.; Schlechter, R.; Ageorges, A.; Arce-Johnson, P.; Tornielli, G.B. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015, 167, 1448–1470. [Google Scholar] [CrossRef]
- An, J.; Liu, Y.; Zhang, X.; Bi, S.; Wang, X.; You, C.; Hao, Y. Dynamic regulation of different light intensity-modulated anthocyanin biosynthesis by BT2-TCP46-MYB1 in apple. J. Exp. Bot. 2020, 71, 3094–3109. [Google Scholar] [CrossRef] [PubMed]
- Ranjith, F.H.; Ariffin, S.H.; Muhialdin, B.J.; Yusof, N.L.; Mohammed, N.K.; Marzlan, A.A.; Hussin, A.S.M. Influence of natural antifungal coatings produced by Lacto-fermented antifungal substances on respiration, quality, antioxidant attributes, and shelf life of mango (Mangifera indica L.). Postharvest Biol. Technol. 2022, 189, 111904. [Google Scholar] [CrossRef]
- Xu, P.Y.; Xu, L.; Xu, H.F.; He, X.W.; He, P.; Chang, Y.S.; Wang, S.; Zheng, W.Y.; Wang, C.Z.; Chen, X.; et al. MdWRKY40is directly promotes anthocyanin accumulation and blocks MdMYB15L, the repressor of MdCBF2, which improves cold tolerance in apple. J. Integr. Agric. 2023, 22, 1704–1719. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Gao, Z.Y.; Hu, M.J.; Pan, Y.G.; Xu, X.B.; Zhang, Z.K. Delay of ripening and senescence in mango fruit by 6-benzylaminopurine is associated with inhibition of ethylene biosynthesis and membrane lipid catabolism. Postharvest Biol. Technol. 2022, 185, 111797. [Google Scholar] [CrossRef]
- Luan, A.P.; Zhang, W.; Yang, M.Z.; Zhong, Z.Q.; Wu, J.; He, Y.H.; He, J.H. Unveiling the molecular mechanism involving anthocyanins in pineapple peel discoloration during fruit maturation. Food Chem. 2023, 412, 135482. [Google Scholar] [CrossRef] [PubMed]
- Zaharah, S.S.; Singh, Z. Mode of action of nitric oxide in inhibiting ethylene biosynthesis and fruit softening during ripening and cool storage of ‘Kensington Pride’ mango. Postharvest Biol. Technol. 2011, 62, 258–266. [Google Scholar] [CrossRef]
- Sudheeran, P.K.; Love, C.; Feygenberg, O.; Maurer, D.; Ovadia, R.; Oren-Shamir, M.; Alkan, N. Induction of red skin and improvement of fruit quality in ‘Kent’, ‘Shelly’ and Maya’ mangoes by preharvest spraying of prohydrojasmon at the orchard. Postharvest Biol. Technol. 2019, 149, 18–26. [Google Scholar] [CrossRef]
- Wang, L.; Yang, S.L.; Ni, N.B.; Teng, Y.W.; Bai, S.L. Advances of anthocyanin synthesis regulated by plant growth regulators in fruit trees. Sci. Hortic. 2023, 307, 111476. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Xu, Y.H.; Li, S.E.; Huan, C.; Jiang, T.J.; Zheng, X.L.; Brecht, J.K. Effects of 1-methylcyclopropene treatment on quality and anthocyanin biosynthesis in plum (Prunus salicina cv. Taoxingli) fruit during storage at a non-chilling temperature. Postharvest Biol. Technol. 2020, 169, 111291. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Pareek, S.; González-Aguilar, G.A.; Domínguez-Avila, J.A. Changes in the activity of proline-metabolising enzymes is associated with increased cultivar-dependent chilling tolerance in mangos, in response to pre-storage melatonin application. Postharvest Biol. Technol. 2021, 182, 11702. [Google Scholar] [CrossRef]
- Chen, X.; Wang, P.; Gu, M.; Hou, B.; Zhang, C.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. Identification of PAL genes related to anthocyanin synthesis in tea plants and its correlation with anthocyanin content. Hortic. Plant J. 2022, 8, 381–394. [Google Scholar] [CrossRef]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 2013, 25, 4135–4149. [Google Scholar] [CrossRef] [PubMed]
- Wako, T.; Kimura, S.; Chikagawa, Y.; Ozeki, Y. Characterization of MYB proteins acting as transcriptional regulatory factors for carrot phenylalanine ammonia-lyase gene (DcPAL3). Plant Biotechnol. 2010, 27, 131–139. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, L.K.; Wang, F.; Espley, R.V.; Ren, F.; Zhao, J.; Ogutu, C.; He, H.; Jiang, Q.; Allan, A.C. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019, 221, 1919–1934. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.M.; Zhou, T.T.; Wang, M.K.; Li, T.T.; Wang, G.B.; Fu, F.F.; Cao, F.L. Systematic investigation and expression profiles of the GbR2R3-MYB transcription factor family in ginkgo (Ginkgo biloba L.). Int. J. Biol. Macromol. 2021, 172, 250–262. [Google Scholar] [CrossRef]
- Colanero, S.; Perata, P.; Gonzali, S. The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 2018, 9, 830. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Ma, D.; Constabel, C.P. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015, 167, 693–710. [Google Scholar] [CrossRef]
- Huang, Y.F.; Vialet, S.; Guiraud, J.L.; Torregrosa, L.; Bertrand, Y.; Cheynier, V.; This, P.; Terrier, N. A negative MYB regulator of proanthocyanidin accumulation was identified through expression quantitative locus mapping in the grape berry. New Phytol. 2014, 201, 795–809. [Google Scholar] [CrossRef]
- Chen, M.M.; Gu, H.; Wang, L.R.; Shao, Y.Z.; Li, R.; Li, W. Exogenous Ethylene Promotes Peel Color Transformation by Regulating the Degradation of Chlorophyll and Synthesis of Anthocyanin in Postharvest Mango Fruit. Front. Nutr. 2022, 20, 911542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pang, X.; Ji, Z.; Jiang, Y. Role of anthocyanin degradation in litchi pericarp browning. Food Chem. 2001, 75, 217–221. [Google Scholar] [CrossRef]
- Shao, Y.Z.; Zeng, J.K.; Tang, H.; Zhou, Y.; Li, W. The chemical treatments combined with antagonistic yeast control anthracnose and maintain the quality of postharvest mango fruit. J. Integr. Agric. 2019, 18, 1159–1169. [Google Scholar] [CrossRef]
- Jiang, Z.C.; Li, R.; Tang, Y.; Cheng, Z.Y.; Qian, M.J.; Li, W.; Shao, Y.Z. Transcriptome Analysis Reveals the Inducing Effect of Bacillus siamensis on Disease Resistance in Postharvest Mango Fruit. Foods 2022, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ma, J.H.; Gu, H.; Jia, W.J.; Shao, Y.Z.; Li, W. 1-Methylcyclopropene counteracts ethylene promotion of fruit softening and roles of MiERF2/8 and MiPG in postharvest mangoes. Front. Plant Sci. 2022, 13, 971050. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.Y.; Zhou, Z.C.; Xiang, X.; Liu, X.; Wang, J.; Hu, Z.R.; Xiang, S.P.; Li, W.; Xiao, Q.Z. Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidase NtRbohE expression. Plant Physiol. 2021, 186, 1706–1720. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aslam, M.M.; Kou, M.; Dou, Y.; Zou, S.; Li, R.; Li, W.; Shao, Y. The Transcription Factor MiMYB8 Suppresses Peel Coloration in Postharvest ‘Guifei’ Mango in Response to High Concentration of Exogenous Ethylene by Negatively Modulating MiPAL1. Int. J. Mol. Sci. 2024, 25, 4841. https://doi.org/10.3390/ijms25094841
Aslam MM, Kou M, Dou Y, Zou S, Li R, Li W, Shao Y. The Transcription Factor MiMYB8 Suppresses Peel Coloration in Postharvest ‘Guifei’ Mango in Response to High Concentration of Exogenous Ethylene by Negatively Modulating MiPAL1. International Journal of Molecular Sciences. 2024; 25(9):4841. https://doi.org/10.3390/ijms25094841
Chicago/Turabian StyleAslam, Muhammad Muzammal, Mingrui Kou, Yaqi Dou, Shicheng Zou, Rui Li, Wen Li, and Yuanzhi Shao. 2024. "The Transcription Factor MiMYB8 Suppresses Peel Coloration in Postharvest ‘Guifei’ Mango in Response to High Concentration of Exogenous Ethylene by Negatively Modulating MiPAL1" International Journal of Molecular Sciences 25, no. 9: 4841. https://doi.org/10.3390/ijms25094841




