Myeloid Cell-Derived IL-1 Signaling Damps Neuregulin-1 from Fibroblasts to Suppress Colitis-Induced Early Repair of the Intestinal Epithelium
Abstract
:1. Introduction
2. Results
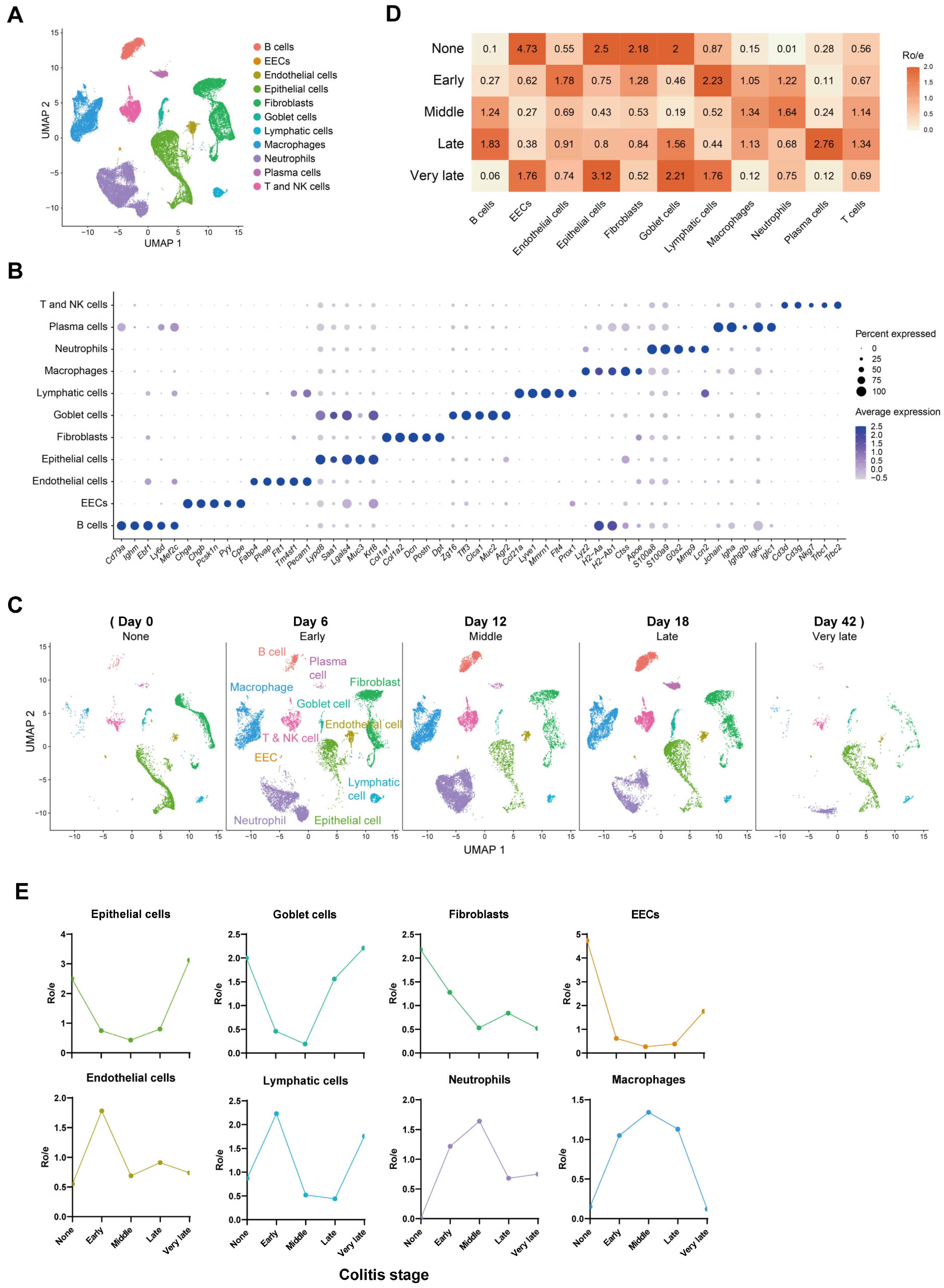
2.1. Composition and Kinetics of Colonic Cells during the Process of Mouse Colitis
2.2. Accumulation of Colonic Nrg1-Expressing Fibroblasts in the Acute Colitis Phase
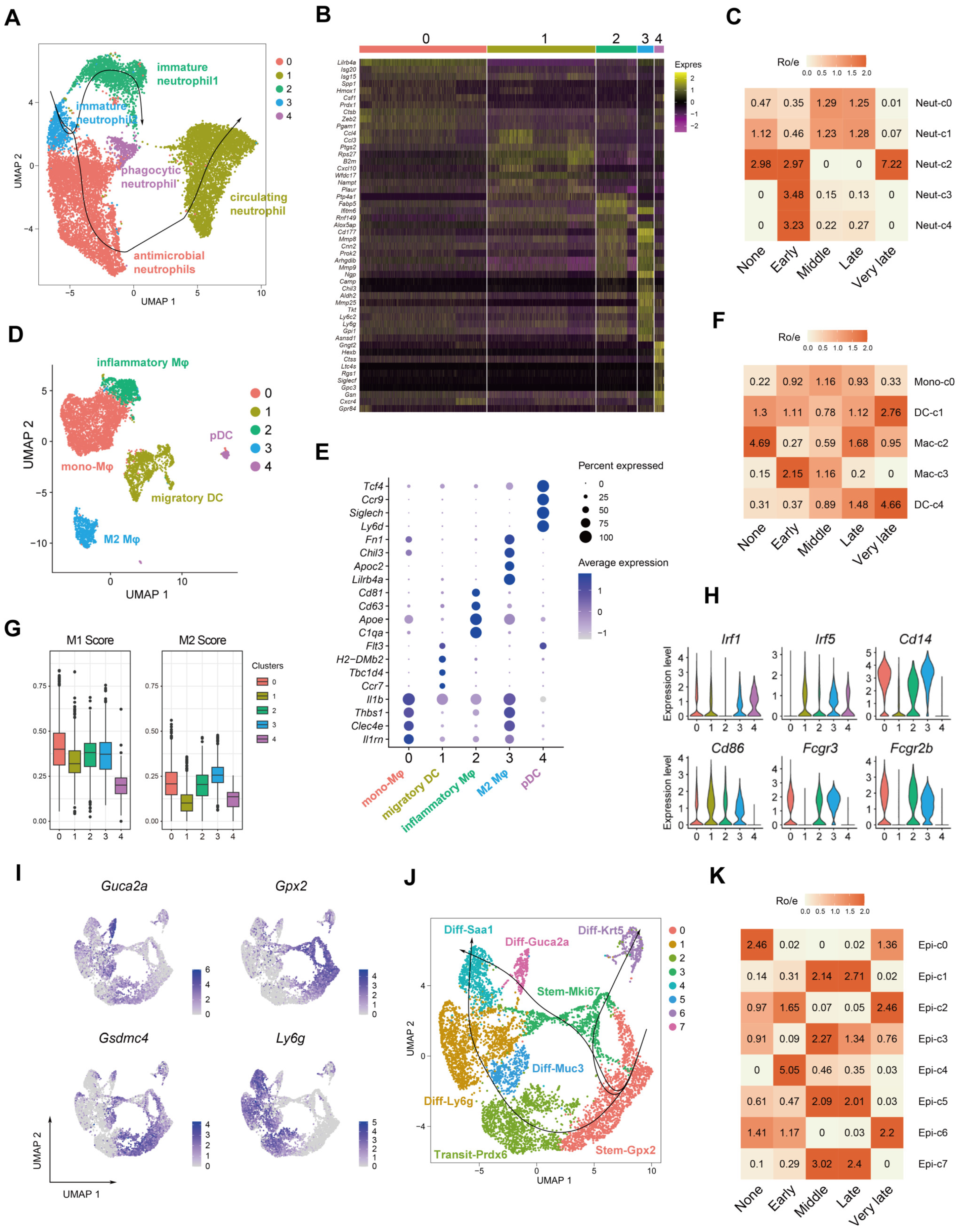
2.3. Dynamics of Myeloid and Epithelial Cells during the Colitis and Recovery Phases
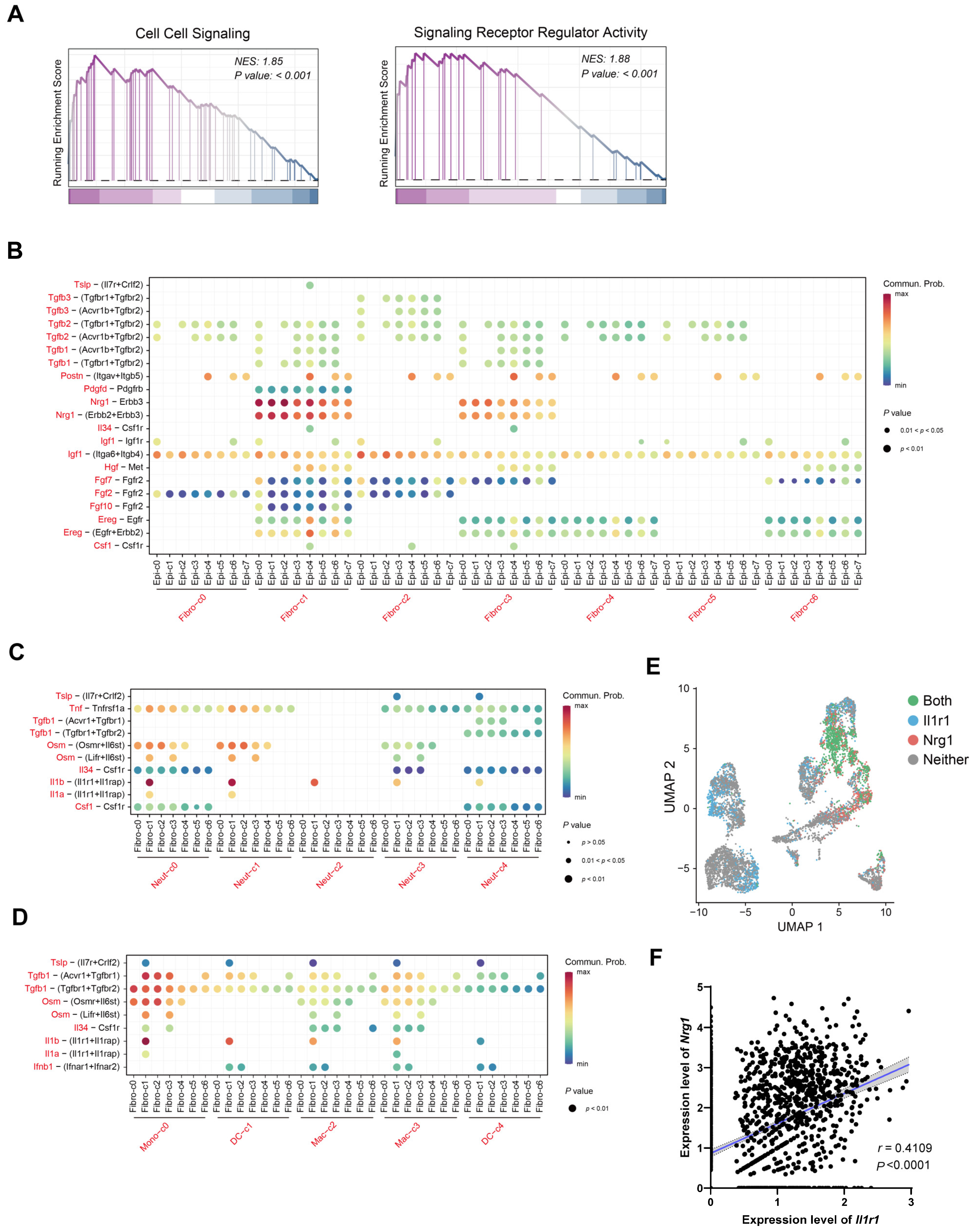
2.4. IL-1 May Influence Nrg1–Erbb3 Interaction between Fibroblasts and Epithelial Cells
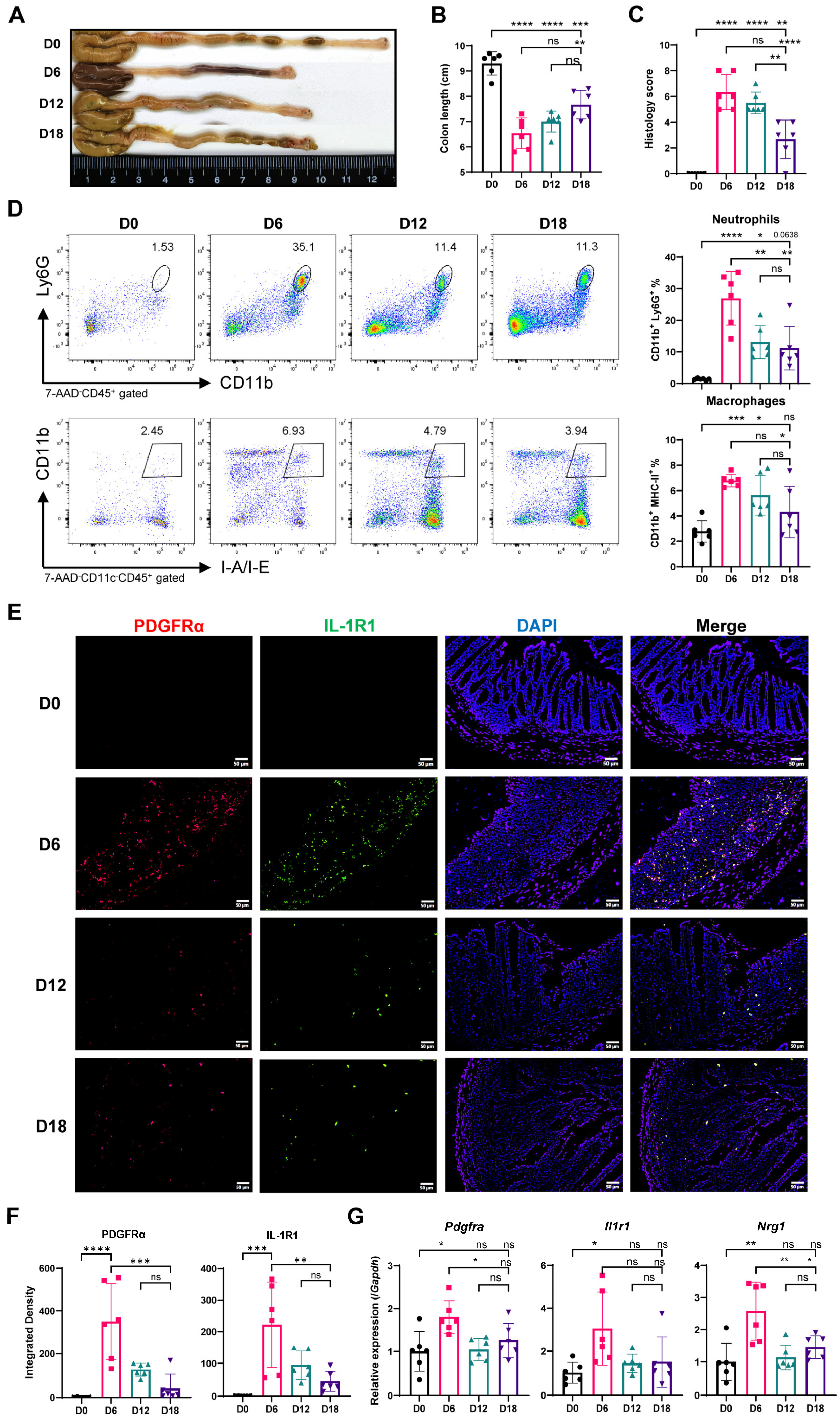
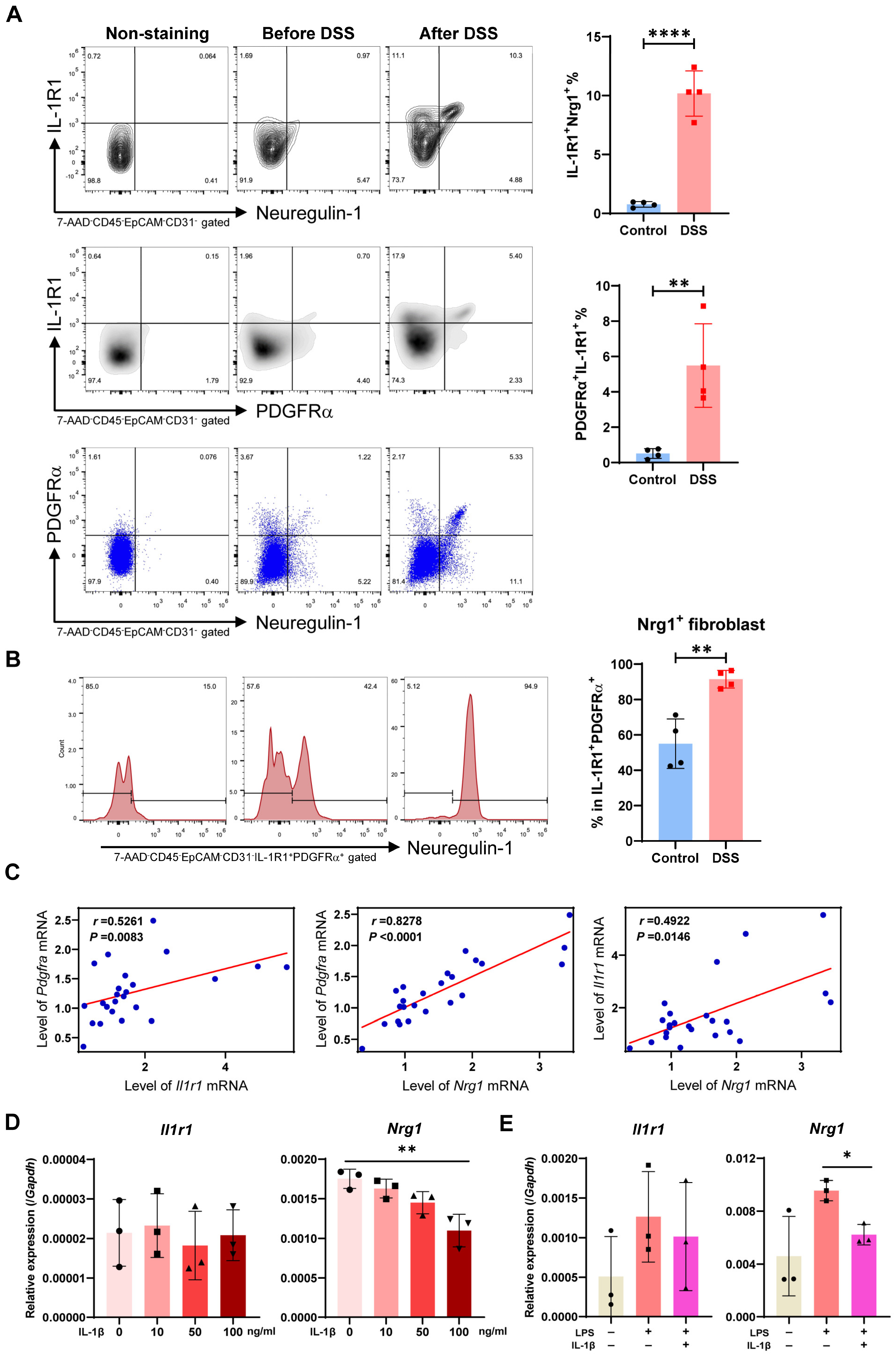
2.5. Myeloid Cell Accumulation and IL-1R and Nrg1 Upregulation in the Process of Colitis-Induced Epithelial Healing
2.6. Most Neuregulin-1-Secreting Fibroblasts Express IL-1R1 and IL-1 Signaling Suppresses Nrg1 Expression under Inflammation Condition
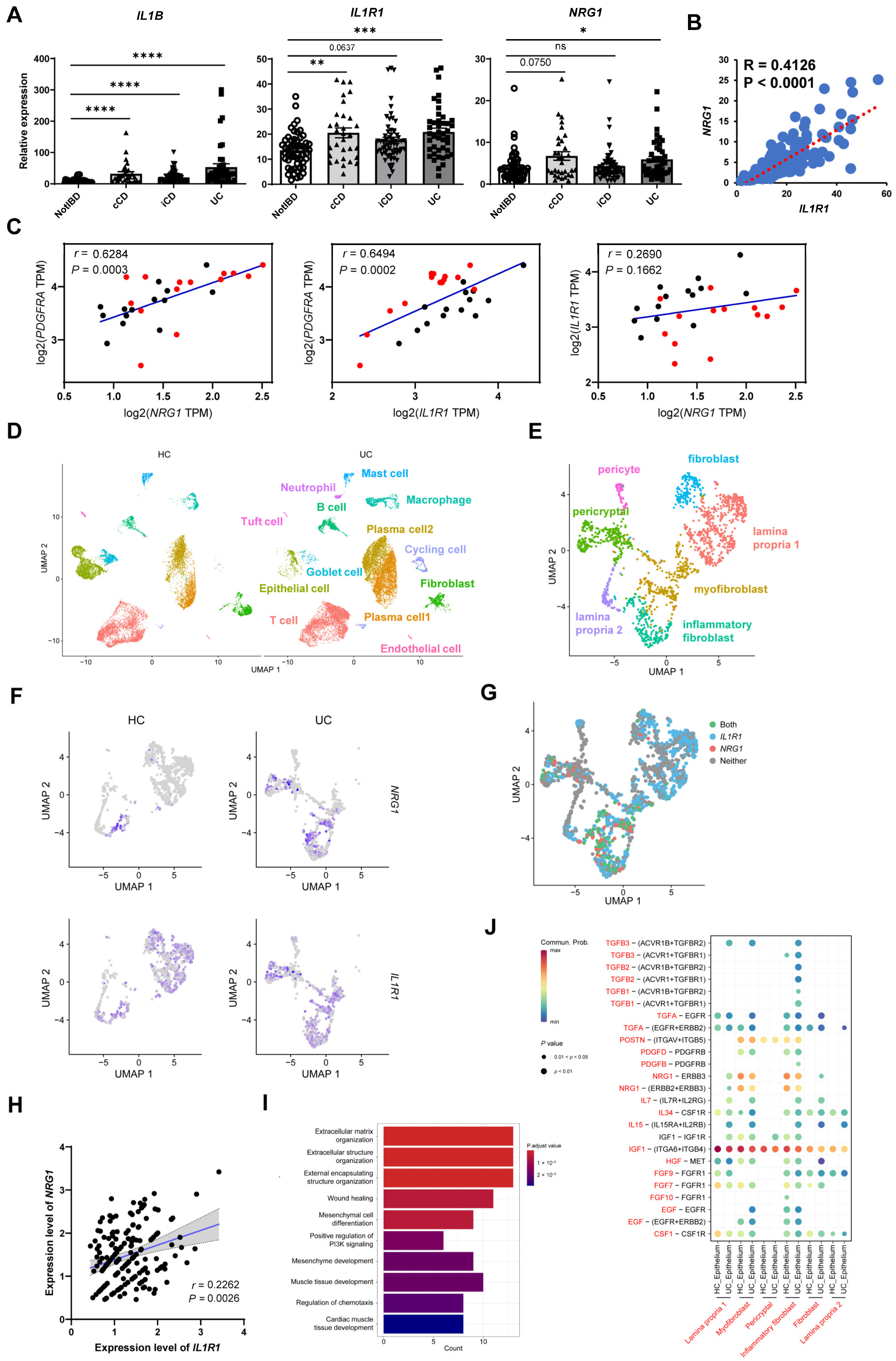
2.7. Co-Expression of NRG1 and IL1R1 Is also Observed in Intestinal Fibroblasts of IBD Patients
3. Discussion
4. Materials and Methods
4.1. Data Collection, Processing, and Cluster Analysis
4.2. Tissue Prevalence of Clusters
4.3. GO Enrichment and GSEA Analysis
4.4. CytoTRACE Analysis
4.5. Pseudotime Trajectory Analysis
4.6. Dorothea Analysis
4.7. Cell–Cell Interaction Analysis
4.8. DSS-Induced Colitis
4.9. Cell Preparation
4.10. Purification and Stimulation of Colonic Fibroblasts
4.11. Culture of Lung Primary Fibroblasts
4.12. Flow Cytometry
4.13. Real-Time RT-PCR
4.14. Histological Analysis
4.15. Immunofluorescence Staining
4.16. Statistical Analysis
5. Conclusions/Summary
- Nrg1 is highly expressed in stem-like fibroblasts that appear early in the mouse colon after inducing colitis with DSS.
- Nrg1 signaling through Erbb3 might mediate communication between colonic fibroblasts and epithelial cells.
- During colitis, there are more myeloid cells releasing IL-1β, along with an increase in fibroblasts that express Nrg1 and IL-1 receptor 1.
- Stimulating mouse fibroblasts with IL-1β reduces Nrg1 expression.
- UC patients also show more NRG1+IL1R1+ colonic fibroblasts and NRG1–ERB interaction between fibroblasts and epithelial cells.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rivière, P.; Li Wai Suen, C.; Chaparro, M.; De Cruz, P.; Spinelli, A.; Laharie, D. Acute Severe Ulcerative Colitis Management: Unanswered Questions and Latest Insights. Lancet Gastroenterol. Hepatol. 2024, 9, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Le Berre, C.; Honap, S.; Peyrin-Biroulet, L. Ulcerative Colitis. Lancet 2023, 402, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Rudbaek, J.J.; Agrawal, M.; Torres, J.; Mehandru, S.; Colombel, J.-F.; Jess, T. Deciphering the Different Phases of Preclinical Inflammatory Bowel Disease. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Dignass, A.U. Mechanisms and Modulation of Intestinal Epithelial Repair. Inflamm. Bowel Dis. 2001, 7, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and Pathogenesis of Inflammatory Bowel Disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Abraham, C.; Dulai, P.S.; Vermeire, S.; Sandborn, W.J. Lessons Learned from Trials Targeting Cytokine Pathways in Patients with Inflammatory Bowel Diseases. Gastroenterology 2017, 152, 374–388.e4. [Google Scholar] [CrossRef]
- Gordon, I.O.; Agrawal, N.; Goldblum, J.R.; Fiocchi, C.; Rieder, F. Fibrosis in Ulcerative Colitis: Mechanisms, Features, and Consequences of a Neglected Problem. Inflamm. Bowel Dis. 2014, 20, 2198–2206. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Zhu, L.; Gorantla, G.; Berdysz, O.; Amici, S.A.; Guerau-de-Arellano, M.; Madalena, K.M.; Lerch, J.K.; Liu, X.; Quan, N. Salient Type 1 Interleukin 1 Receptor Expression in Peripheral Non-Immune Cells. Sci. Rep. 2018, 8, 723. [Google Scholar] [CrossRef] [PubMed]
- Birchmeier, C.; Nave, K.-A. Neuregulin-1, a Key Axonal Signal That Drives Schwann Cell Growth and Differentiation. Glia 2008, 56, 1491–1497. [Google Scholar] [CrossRef]
- Britsch, S. The Neuregulin-I/ErbB Signaling System in Development and Disease. Adv. Anat. Embryol. Cell Biol. 2007, 190, 1–65. [Google Scholar]
- Mei, L.; Xiong, W.-C. Neuregulin 1 in Neural Development, Synaptic Plasticity and Schizophrenia. Nat. Rev. Neurosci. 2008, 9, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Farkas, J.E.; Freitas, P.D.; Bryant, D.M.; Whited, J.L.; Monaghan, J.R. Neuregulin-1 Signaling Is Essential for Nerve-Dependent Axolotl Limb Regeneration. Development 2016, 143, 2724–2731. [Google Scholar] [CrossRef] [PubMed]
- Bartus, K.; Galino, J.; James, N.D.; Hernandez-Miranda, L.R.; Dawes, J.M.; Fricker, F.R.; Garratt, A.N.; McMahon, S.B.; Ramer, M.S.; Birchmeier, C.; et al. Neuregulin-1 Controls an Endogenous Repair Mechanism after Spinal Cord Injury. Brain 2016, 139, 1394–1416. [Google Scholar] [CrossRef]
- Rupert, C.E.; Coulombe, K.L. The Roles of Neuregulin-1 in Cardiac Development, Homeostasis, and Disease. Biomark Insights 2015, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Trigo, A.; Corraliza, A.M.; Veny, M.; Dotti, I.; Melón-Ardanaz, E.; Rill, A.; Crowell, H.L.; Corbí, Á.; Gudiño, V.; Esteller, M.; et al. Macrophage and Neutrophil Heterogeneity at Single-Cell Spatial Resolution in Human Inflammatory Bowel Disease. Nat. Commun. 2023, 14, 4506. [Google Scholar] [CrossRef] [PubMed]
- Jardé, T.; Chan, W.H.; Rossello, F.J.; Kaur Kahlon, T.; Theocharous, M.; Kurian Arackal, T.; Flores, T.; Giraud, M.; Richards, E.; Chan, E.; et al. Mesenchymal Niche-Derived Neuregulin-1 Drives Intestinal Stem Cell Proliferation and Regeneration of Damaged Epithelium. Cell Stem Cell 2020, 27, 646–662.e7. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.B.; Storm, E.E.; Kapoor, V.N.; Chavarria-Smith, J.; Lin, D.L.; Wang, L.; Li, Y.; Kljavin, N.; Ota, N.; Bainbridge, T.W.; et al. IL-1R1–Dependent Signaling Coordinates Epithelial Regeneration in Response to Intestinal Damage. Sci. Immunol. 2021, 6, eabe8856. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, D.; Italiani, P.; Weil, S.; Martin, M.U. The Family of the Interleukin-1 Receptors. Immunol. Rev. 2018, 281, 197–232. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA Sequencing: The Teenage Years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Liu, C.Y.; Girish, N.; Gomez, M.L.; Kalski, M.; Bernard, J.K.; Simons, B.D.; Polk, D.B. Wound-Healing Plasticity Enables Clonal Expansion of Founder Progenitor Cells in Colitis. Dev. Cell 2023, 58, 2309–2325.e7. [Google Scholar] [CrossRef]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [PubMed]
- Buechler, M.B.; Pradhan, R.N.; Krishnamurty, A.T.; Cox, C.; Calviello, A.K.; Wang, A.W.; Yang, Y.A.; Tam, L.; Caothien, R.; Roose-Girma, M.; et al. Cross-Tissue Organization of the Fibroblast Lineage. Nature 2021, 593, 575–579. [Google Scholar] [CrossRef]
- Jasso, G.J.; Jaiswal, A.; Varma, M.; Laszewski, T.; Grauel, A.; Omar, A.; Silva, N.; Dranoff, G.; Porter, J.A.; Mansfield, K.; et al. Colon Stroma Mediates an Inflammation-Driven Fibroblastic Response Controlling Matrix Remodeling and Healing. PLoS Biol. 2022, 20, e3001532. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Goudarzi, K.M.; Abedi, S.; Pieber, M.; Sjöberg, E.; Behnan, J.; Zhang, X.-M.; Harris, R.A.; Bartek, J.; Lindström, M.S.; et al. SFRP2 Induces a Mesenchymal Subtype Transition by Suppression of SOX2 in Glioblastoma. Oncogene 2021, 40, 5066–5080. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Nguyen, Q.T.; Li, J.; Zhao, S.; Christensen, S.M.; West, G.A.; Chandra, J.; Gordon, I.O.; Lin, S.; Wang, J.; et al. Stricturing Crohn’s Disease Single-Cell RNA Sequencing Reveals Fibroblast Heterogeneity and Intercellular Interactions. Gastroenterology 2023, 165, 1180–1196. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the Path to Cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Wang, P.; Su, J.; Wang, J.; Xie, Y.; Chen, W.; Zhong, J.; Wang, Y. NRF1 Promotes Primordial Germ Cell Development, Proliferation and Survival. Cell Prolif. 2024, 57, e13533. [Google Scholar] [CrossRef] [PubMed]
- Ebright, R.Y.; Zachariah, M.A.; Micalizzi, D.S.; Wittner, B.S.; Niederhoffer, K.L.; Nieman, L.T.; Chirn, B.; Wiley, D.F.; Wesley, B.; Shaw, B.; et al. HIF1A Signaling Selectively Supports Proliferation of Breast Cancer in the Brain. Nat. Commun. 2020, 11, 6311. [Google Scholar] [CrossRef]
- Garcia-Rendueles, M.E.R.; Krishnamoorthy, G.; Saqcena, M.; Acuña-Ruiz, A.; Revilla, G.; de Stanchina, E.; Knauf, J.A.; Lester, R.; Xu, B.; Ghossein, R.A.; et al. Yap Governs a Lineage-Specific Neuregulin1 Pathway-Driven Adaptive Resistance to RAF Kinase Inhibitors. Mol. Cancer 2022, 21, 213. [Google Scholar] [CrossRef]
- Artap, S.; Manderfield, L.J.; Smith, C.L.; Poleshko, A.; Aghajanian, H.; See, K.; Li, L.; Jain, R.; Epstein, J.A. Endocardial Hippo Signaling Regulates Myocardial Growth and Cardiogenesis. Dev. Biol. 2018, 440, 22–30. [Google Scholar] [CrossRef]
- Magoro, T.; Dandekar, A.; Jennelle, L.T.; Bajaj, R.; Lipkowitz, G.; Angelucci, A.R.; Bessong, P.O.; Hahn, Y.S. IL-1β/TNF-α/IL-6 Inflammatory Cytokines Promote STAT1-Dependent Induction of CH25H in Zika Virus-Infected Human Macrophages. J. Biol. Chem. 2019, 294, 14591–14602. [Google Scholar] [CrossRef] [PubMed]
- Whitley, S.K.; Balasubramani, A.; Zindl, C.L.; Sen, R.; Shibata, Y.; Crawford, G.E.; Weathington, N.M.; Hatton, R.D.; Weaver, C.T. IL-1R Signaling Promotes STAT3 and NF-κB Factor Recruitment to Distal Cis-Regulatory Elements That Regulate Il17a/f Transcription. J. Biol. Chem. 2018, 293, 15790–15800. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Shi, Q.; Wu, P.; Zhang, X.; Kambara, H.; Su, J.; Yu, H.; Park, S.-Y.; Guo, R.; Ren, Q.; et al. Single-Cell Transcriptome Profiling Reveals Neutrophil Heterogeneity in Homeostasis and Infection. Nat. Immunol. 2020, 21, 1119–1133. [Google Scholar] [CrossRef] [PubMed]
- Vafadarnejad, E.; Rizzo, G.; Krampert, L.; Arampatzi, P.; Arias-Loza, A.-P.; Nazzal, Y.; Rizakou, A.; Knochenhauer, T.; Bandi, S.R.; Nugroho, V.A.; et al. Dynamics of Cardiac Neutrophil Diversity in Murine Myocardial Infarction. Circ. Res. 2020, 127, e232–e249. [Google Scholar] [CrossRef] [PubMed]
- Hilligan, K.L.; Ronchese, F. Antigen Presentation by Dendritic Cells and Their Instruction of CD4+ T Helper Cell Responses. Cell Mol. Immunol. 2020, 17, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Gallego, C.; Vétillard, M.; Calmette, J.; Roriz, M.; Marin-Esteban, V.; Evrard, M.; Aknin, M.-L.; Pionnier, N.; Lefrançois, M.; Mercier-Nomé, F.; et al. CXCR4 Signaling Controls Dendritic Cell Location and Activation at Steady State and in Inflammation. Blood 2021, 137, 2770–2784. [Google Scholar] [CrossRef] [PubMed]
- Swiecki, M.; Wang, Y.; Riboldi, E.; Kim, A.H.J.; Dzutsev, A.; Gilfillan, S.; Vermi, W.; Ruedl, C.; Trinchieri, G.; Colonna, M. Cell Depletion in Mice That Express Diphtheria Toxin Receptor under the Control of SiglecH Encompasses More than Plasmacytoid Dendritic Cells. J. Immunol. 2014, 192, 4409–4416. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P.; et al. A Pan-Cancer Single-Cell Transcriptional Atlas of Tumor Infiltrating Myeloid Cells. Cell 2021, 184, 792–809.e23. [Google Scholar] [CrossRef] [PubMed]
- Girish, N.; Liu, C.Y.; Gadeock, S.; Gomez, M.L.; Huang, Y.; Sharifkhodaei, Z.; Washington, M.K.; Polk, D.B. Persistence of Lgr5+ Colonic Epithelial Stem Cells in Mouse Models of Inflammatory Bowel Disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G308–G324. [Google Scholar] [CrossRef]
- Ohara, T.E.; Colonna, M.; Stappenbeck, T.S. Adaptive Differentiation Promotes Intestinal Villus Recovery. Dev. Cell 2022, 57, 166–179.e6. [Google Scholar] [CrossRef]
- Lee, P.; Gund, R.; Dutta, A.; Pincha, N.; Rana, I.; Ghosh, S.; Witherden, D.; Kandyba, E.; MacLeod, A.; Kobielak, K.; et al. Stimulation of Hair Follicle Stem Cell Proliferation through an IL-1 Dependent Activation of γδT-Cells. eLife 2017, 6, e28875. [Google Scholar] [CrossRef]
- Katsura, H.; Kobayashi, Y.; Tata, P.R.; Hogan, B.L.M. IL-1 and TNFα Contribute to the Inflammatory Niche to Enhance Alveolar Regeneration. Stem Cell Rep. 2019, 12, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Patankar, J.V.; Becker, C. Cell Death in the Gut Epithelium and Implications for Chronic Inflammation. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 543–556. [Google Scholar] [CrossRef]
- Zhu, G.; Hu, J.; Xi, R. The Cellular Niche for Intestinal Stem Cells: A Team Effort. Cell Regen. 2021, 10, 1. [Google Scholar] [CrossRef]
- Talbott, H.E.; Mascharak, S.; Griffin, M.; Wan, D.C.; Longaker, M.T. Wound Healing, Fibroblast Heterogeneity, and Fibrosis. Cell Stem Cell 2022, 29, 1161–1180. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Pohin, M.; Jackson, M.A.; Korsunsky, I.; Bullers, S.J.; Rue-Albrecht, K.; Christoforidou, Z.; Sathananthan, D.; Thomas, T.; Ravindran, R.; et al. IL-1-Driven Stromal-Neutrophil Interactions Define a Subset of Patients with Inflammatory Bowel Disease That Does Not Respond to Therapies. Nat. Med. 2021, 27, 1970–1981. [Google Scholar] [CrossRef]
- Koncina, E.; Nurmik, M.; Pozdeev, V.I.; Gilson, C.; Tsenkova, M.; Begaj, R.; Stang, S.; Gaigneaux, A.; Weindorfer, C.; Rodriguez, F.; et al. IL1R1+ Cancer-Associated Fibroblasts Drive Tumor Development and Immunosuppression in Colorectal Cancer. Nat. Commun. 2023, 14, 4251. [Google Scholar] [CrossRef]
- Nicolas, A.M.; Pesic, M.; Engel, E.; Ziegler, P.K.; Diefenhardt, M.; Kennel, K.B.; Buettner, F.; Conche, C.; Petrocelli, V.; Elwakeel, E.; et al. Inflammatory Fibroblasts Mediate Resistance to Neoadjuvant Therapy in Rectal Cancer. Cancer Cell 2022, 40, 168–184.e13. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef]
- Fenton, C.G.; Taman, H.; Florholmen, J.; Sørbye, S.W.; Paulssen, R.H. Transcriptional Signatures That Define Ulcerative Colitis in Remission. Inflamm. Bowel Dis. 2021, 27, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage Tracking Reveals Dynamic Relationships of T Cells in Colorectal Cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Gulati, G.S.; Sikandar, S.S.; Wesche, D.J.; Manjunath, A.; Bharadwaj, A.; Berger, M.J.; Ilagan, F.; Kuo, A.H.; Hsieh, R.W.; Cai, S.; et al. Single-Cell Transcriptional Diversity Is a Hallmark of Developmental Potential. Science 2020, 367, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed Graph Embedding Resolves Complex Single-Cell Trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and Integration of Resources for the Estimation of Human Transcription Factor Activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.-H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and Analysis of Cell-Cell Communication Using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Cooper, H.S.; Murthy, S.N.; Shah, R.S.; Sedergran, D.J. Clinicopathologic Study of Dextran Sulfate Sodium Experimental Murine Colitis. Lab. Investig. 1993, 69, 238–249. [Google Scholar] [PubMed]
- Arstila, T.; Arstila, T.P.; Calbo, S.; Selz, F.; Malassis-Seris, M.; Vassalli, P.; Kourilsky, P.; Guy-Grand, D. Identical T Cell Clones Are Located within the Mouse Gut Epithelium and Lamina Propia and Circulate in the Thoracic Duct Lymph. J. Exp. Med. 2000, 191, 823–834. [Google Scholar] [CrossRef]
- Obermeier, F.; Dunger, N.; Deml, L.; Herfarth, H.; Schölmerich, J.; Falk, W. CpG Motifs of Bacterial DNA Exacerbate Colitis of Dextran Sulfate Sodium-Treated Mice. Eur. J. Immunol. 2002, 32, 2084–2092. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, D.; Xu, S.; Ji, K.; Tang, C. Myeloid Cell-Derived IL-1 Signaling Damps Neuregulin-1 from Fibroblasts to Suppress Colitis-Induced Early Repair of the Intestinal Epithelium. Int. J. Mol. Sci. 2024, 25, 4469. https://doi.org/10.3390/ijms25084469
Qiu D, Xu S, Ji K, Tang C. Myeloid Cell-Derived IL-1 Signaling Damps Neuregulin-1 from Fibroblasts to Suppress Colitis-Induced Early Repair of the Intestinal Epithelium. International Journal of Molecular Sciences. 2024; 25(8):4469. https://doi.org/10.3390/ijms25084469
Chicago/Turabian StyleQiu, Ding, Shaoting Xu, Kaile Ji, and Ce Tang. 2024. "Myeloid Cell-Derived IL-1 Signaling Damps Neuregulin-1 from Fibroblasts to Suppress Colitis-Induced Early Repair of the Intestinal Epithelium" International Journal of Molecular Sciences 25, no. 8: 4469. https://doi.org/10.3390/ijms25084469




